Protein vs. DNA: A Comparative Analysis of Food Allergen Detection Methods for Biomedical Research
This article provides a comprehensive comparative analysis of protein-based and DNA-based methods for food allergen detection, a critical area for food safety and public health.
Protein vs. DNA: A Comparative Analysis of Food Allergen Detection Methods for Biomedical Research
Abstract
This article provides a comprehensive comparative analysis of protein-based and DNA-based methods for food allergen detection, a critical area for food safety and public health. Tailored for researchers, scientists, and drug development professionals, it explores the fundamental principles, advantages, and limitations of immunological assays (e.g., ELISA, lateral flow) and mass spectrometry versus nucleic acid techniques (e.g., PCR, LAMP, real-time PCR). The scope extends to methodological applications, troubleshooting for complex food matrices, optimization strategies to overcome technological limitations, and a rigorous validation framework for method selection. The analysis synthesizes current research trends, including biosensors and high-throughput technologies, to guide future innovations in allergen detection and clinical diagnostics.
Core Principles and the Evolving Landscape of Food Allergen Detection
Food allergy has emerged as a critical public health challenge worldwide, with documented prevalence increases across multiple continents. In China, infant food allergy incidence rose from 7.7% in 2009 to 11.1% in 2019, while similar upward trends are observed in the United States, United Kingdom, and Europe [1] [2]. This rising prevalence, coupled with the potential severity of allergic reactions including life-threatening anaphylaxis, has intensified the focus on reliable allergen detection methods for protective public health measures [1] [3].
With no specific treatment currently available for food allergies, strict avoidance of allergenic foods remains the primary preventive strategy [1]. This approach depends entirely on accurate food labeling, which in turn relies on robust detection methodologies to verify allergen presence [4]. Regulatory frameworks globally have responded by implementing mandatory allergen labeling requirements for major allergens, with the European Union's regulation (EU) No 1169/2011 expanding its list to 14 key allergenic foods including celery, sesame, mustard, lupin, and mollusks alongside traditional allergens [5].
This article provides a comparative analysis of two fundamental allergen detection approaches: protein-based methods (including ELISA and mass spectrometry) and DNA-based methods (primarily PCR-based techniques), examining their performance characteristics, limitations, and appropriate applications within the context of rising public health demands and regulatory compliance needs.
Methodological Approaches: Fundamental Principles and Techniques
Protein-Based Detection Methods
Protein-based methods directly target the allergenic proteins themselves and represent the current mainstream approach for food allergen detection [1]. These techniques include:
Enzyme-Linked Immunosorbent Assay (ELISA): This immunological method uses antibodies specific to allergenic proteins and is characterized by high sensitivity, strong specificity, and multi-detection capability [1]. ELISA is recognized as the gold standard for routine allergen screening due to its cost-effectiveness and reliability across various food matrices [4]. The Codex Alimentarius Commission has adopted ELISA as the official test for gluten allergens, setting a threshold of 20 mg/kg for gluten in foods [1].
Mass Spectrometry (MS): LC-MS/MS methods detect and quantify specific allergen proteins by targeting proteotypic peptides unique to each allergen, offering high specificity and the ability to detect multiple allergens simultaneously (multiplexing) [6]. Mass spectrometry is particularly valuable for complex matrices and provides new levels of precision compared to other methods [6].
DNA-Based Detection Methods
DNA-based methods, primarily polymerase chain reaction (PCR) techniques, target allergen-encoding genes rather than the proteins themselves [1]. These methods offer particular advantages for detecting allergens in highly processed foods where proteins may be denatured but DNA remains stable [5]. Key DNA-based approaches include:
Quantitative PCR (qPCR): Enables quantification of allergen DNA through real-time amplification monitoring, with detection sensitivity down to 1 ppm of spiked protein in various food matrices [5].
Loop-Mediated Isothermal Amplification (LAMP): Provides rapid detection without requiring thermal cycling equipment, making it suitable for field applications [1].
Digital PCR and Saltatory Rolling Circle Amplification: Emerging techniques offering enhanced sensitivity and specificity for challenging applications [1].
Comparative Performance Analysis: Experimental Data and Validation
Sensitivity and Detection Limits
Table 1: Sensitivity Comparison Between Protein-Based and DNA-Based Methods
| Method | Detection Limit | Representative Allergens Detected | Experimental Matrix |
|---|---|---|---|
| ELISA | Parts per million (ppm) levels | Gluten, milk, egg, peanut, soy [4] | Processed foods, raw ingredients [4] |
| Mass Spectrometry | As low as 0.01 ng/mL [6] | Peanut (Ara h 3, Ara h 6), milk (Bos d 5), egg (Gal d 1, Gal d 2) [6] | Complex food matrices [6] |
| qPCR | 1 ppm spiked protein [5] | Celery, wheat, maize [5] [7] | Five AOAC food matrix groups [5] |
| Novel PCR Methods | Detection after 220°C/60min baking [7] | Wheat HMW-GS, LMW-GS, maize Zea m 14, Zea m 8, zein [7] | Baked goods, processed foods [7] |
Matrix Effects and Processing Impacts
Food matrix composition significantly influences method performance. A 2024 comparative study of DNA-based celery detection kits found a clear matrix effect across five product groups ((plant-based) meat products, snacks, sauces, dried herbs and spices, and smoothies), despite all kits performing according to specifications [5]. Quantitative performance proved challenging in all food product groups using DNA-based quantification methods [5].
Processing conditions, particularly thermal treatment, dramatically affect detection capability. Protein-based methods may fail when allergenic proteins are denatured by high temperatures, while DNA-based methods face challenges from DNA fragmentation. Research demonstrates that DNA integrity decreases with increasing processing temperature and time, with optimal PCR amplification requiring shorter amplicons (∼200–300 bp) in processed foods [7].
Table 2: Impact of Food Processing on Allergen Detection Methods
| Processing Condition | Effect on Protein-Based Methods | Effect on DNA-Based Methods | Experimental Evidence |
|---|---|---|---|
| Thermal Processing (Baking 180-220°C) | Protein denaturation reduces antibody recognition [5] | DNA degradation requires shorter amplicons (200-300 bp) [7] | Wheat/maize DNA detectable after 60min at 220°C with optimized primers [7] |
| Highly Processed Matrices | Potential epitope destruction limits antibody binding [5] | Higher DNA stability maintains detectability [5] | DNA methods effective for thermally processed foods where proteins denatured [5] |
| Complex Matrices (Sauces, Spices) | Component interference with antibody binding [5] | PCR inhibitors affect amplification efficiency [5] | Clear matrix effect observed in celery detection across 5 product groups [5] |
Quantitative Performance and Regulatory Application
Quantification represents a particular challenge for DNA-based methods. The 2024 celery detection study noted that conversion of DNA results leads to overestimation of the amount of celery, indicating that DNA-based quantification requires optimization for reliable risk management decisions [5]. This limitation is significant in regulatory contexts where precise threshold adherence is mandatory.
Protein-based methods, particularly ELISA, are widely adopted in regulatory frameworks due to their reliable quantification capabilities. Japan recognizes both ELISA and PCR as official testing methods with a defined food allergen threshold of 10 μg/g, while the Codex Alimentarius specifies 20 mg/kg for gluten based on ELISA testing [1].
Method Selection Framework: Technical Considerations
Experimental Workflows
The fundamental methodological differences between protein-based and DNA-based approaches necessitate distinct experimental workflows:
Decision Framework for Method Selection
The choice between protein-based and DNA-based methods depends on multiple experimental factors:
Research Reagent Solutions: Essential Materials and Tools
Table 3: Essential Research Reagents for Allergen Detection Methods
| Reagent/Material | Function | Application Examples |
|---|---|---|
| Allergen-Specific Antibodies | Binds specifically to target allergenic proteins for detection | ELISA for gluten, peanut, milk proteins [4] |
| CTAB Buffer | Cell lysis and DNA stabilization during extraction | DNA extraction from celery, wheat, maize [5] [7] |
| Proteinase K | Protein digestion for DNA purification or protein analysis | Sample preparation in DNA extraction and MS analysis [5] |
| TaqMan Probes & Primers | Sequence-specific DNA detection in qPCR | Celery detection with Cel-MDH primers/probe [5] |
| Trypsin | Protein digestion for mass spectrometric analysis | Generation of proteotypic peptides for LC-MS/MS [6] |
| Reference Allergen Standards | Quantification and method calibration | Certified reference materials for allergen quantification [5] |
Emerging technologies are poised to transform the allergen detection landscape. Biosensors showing promise for rapid, on-site detection are being developed with high sensitivity and specificity [1]. AI-enhanced methods including hyperspectral imaging and FTIR spectroscopy enable non-destructive, real-time allergen detection without compromising food integrity [6]. Additionally, multiplexed platforms capable of simultaneously detecting multiple allergens address the growing need for comprehensive screening approaches [6].
The future of allergen detection lies in integrating complementary methodologies to leverage their respective strengths while mitigating limitations. As detection technologies evolve alongside increasing regulatory scrutiny and public health demands, the development of accurate, reliable, and accessible allergen detection platforms remains crucial for protecting susceptible populations worldwide.
For researchers and food safety professionals, selecting the optimal allergen detection method is critical for accurate risk assessment and regulatory compliance. The core challenge lies in choosing between two fundamental approaches: protein-based methods, which directly detect the allergenic molecules themselves, and DNA-based methods, which identify the genetic material encoding these proteins. Protein-based techniques, including Enzyme-Linked Immunosorbent Assay (ELISA) and mass spectrometry, are the established standard for directly measuring allergenic proteins, the very compounds that trigger immune responses in sensitive individuals [1] [8]. However, the evolving complexity of food matrices and processing techniques demands a thorough comparative analysis to guide methodological selection. This guide provides an objective, data-driven comparison of protein-based detection with DNA-based alternatives, focusing on performance characteristics, experimental protocols, and suitability for different research and quality control scenarios.
Performance Comparison: Protein-Based vs. DNA-Based Methods
The choice between protein-based and DNA-based detection methods involves a careful trade-off between directness, sensitivity, and robustness to food processing. The table below summarizes the core characteristics of each approach.
Table 1: Core Characteristics of Allergen Detection Methodologies
| Feature | Protein-Based Methods | DNA-Based Methods |
|---|---|---|
| Target Analyte | Allergenic proteins (e.g., Gly m 5 in soybean, Tri a 19 in wheat) [9] | DNA sequences encoding allergenic proteins or species-specific genes [1] [9] |
| Directness | Directly measures the causative agent of the allergic reaction [8] | Indirect; infers allergen presence via genetic material [8] |
| Key Advantage | High biological relevance; can correlate with allergenic potential [8] | Superior stability of DNA in processed foods; high sensitivity and specificity [10] [1] |
| Key Limitation | Susceptible to protein denaturation and epitope masking from processing, leading to potential false negatives [9] [8] | Does not directly quantify the allergenic protein; results can be influenced by gene copy number variation [8] |
Beyond these fundamental differences, the practical performance of these methods varies significantly in terms of sensitivity, throughput, and operational requirements.
Table 2: Performance and Operational Comparison
| Performance Metric | Protein-Based Methods | DNA-Based Methods |
|---|---|---|
| Typical Sensitivity | ppm (mg/kg) to low ppb (μg/kg) levels [1] [11] | Can detect down to 1-10 pg/μL of genomic DNA or 0.1% (w/w) of allergenic material [12] [13] [11] |
| Multiplexing Capability | Possible with advanced techniques like multiplex immunoassays or mass spectrometry [8] | Possible with real-time PCR arrays or isothermal amplification, but often requires multiple parallel reactions [9] |
| Throughput | High for ELISA; lower for MS-based methods | High for PCR and isothermal amplification |
| Cost & Expertise | ELISA is relatively low-cost; MS requires expensive instrumentation and specialized expertise [9] | Requires thermocyclers (for PCR) or precise heating blocks; expertise in molecular biology needed |
| Standardization | Well-established as official methods (e.g., ELISA for gluten by Codex Alimentarius) [1] | Recognized as official methods in several countries (e.g., Germany, Japan) [1] |
Experimental Data and Validation
Quantitative Data from Comparative Studies
Independent studies and proficiency testing schemes provide critical insights into the real-world performance of these methods. For instance, a comparative assessment of commercial DNA test kits for celery allergen detection highlighted that while they performed according to specifications, a clear matrix effect was observed across different food product groups, and quantitative performance remained challenging [12]. This underscores that claimed sensitivity can be significantly impacted by the food matrix.
The UK Food Standards Agency's extensive literature review on allergen testing methodologies further confirms that the performance of commercial ELISA and PCR kits can vary, with limits of detection (LOD) and quantitation (LOQ) being periodically updated by manufacturers. This makes direct, time-independent kit-to-kit comparisons difficult [11].
Impact of Food Processing
Food processing is a major factor influencing method selection. Thermal and non-thermal processing can induce protein denaturation, aggregation, and chemical modification (e.g., Maillard reaction), which can mask or destroy antibody-binding epitopes [8]. This can lead to false negatives in protein-based assays like ELISA [9].
In contrast, DNA is generally more stable during processing. However, it is not immune to degradation. A study on wheat and maize demonstrated that while genomic DNA degrades with high-temperature baking (e.g., 220°C for 60 minutes), PCR detection remains possible by targeting short, stable DNA fragments (∼200–300 bp) [10]. This principle is leveraged in novel isothermal amplification methods. For example, the duplex Proofman-LMTIA method developed for soybean and wheat allergen detection achieved a sensitivity of 10 pg/μL genomic DNA and was effective in processed commercial products, whereas a standard LAMP method produced false positives [9].
Experimental Protocols in Practice
Key Workflows
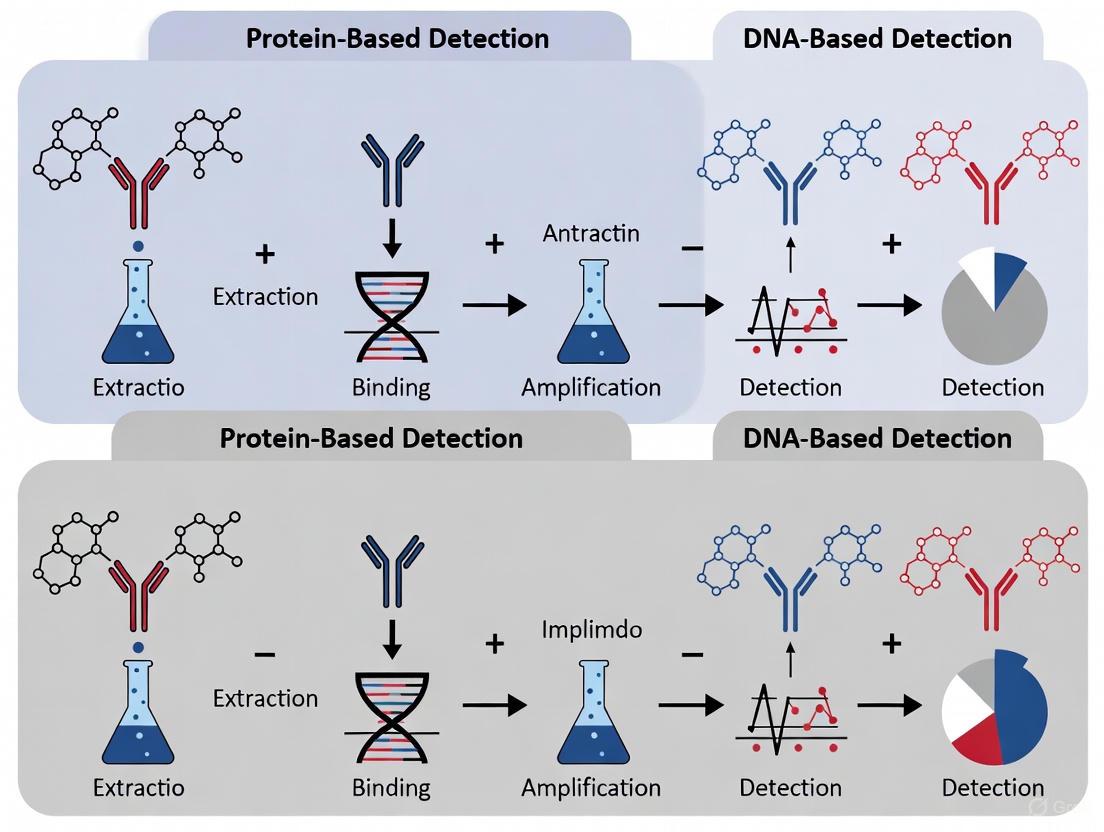
The fundamental workflows for protein-based and DNA-based detection differ significantly, from sample preparation to final analysis. The following diagram outlines the key steps for the primary methodologies in each category.
Detailed Methodological Protocols
Protein-Based Protocol: ELISA for Allergen Quantification
The ELISA is a cornerstone protein-based method. A typical sandwich ELISA protocol involves:
- Coating: A multi-well plate is coated with a capture antibody specific to the target allergen.
- Blocking: The plate is blocked with a protein buffer (e.g., BSA or casein) to prevent non-specific binding.
- Incubation with Sample: The extracted food sample and a series of allergen standards of known concentration are added to separate wells.
- Incubation with Detection Antibody: A second antibody, specific to a different epitope on the same allergen and conjugated to an enzyme (e.g., Horseradish Peroxidase, HRP), is added.
- Signal Development: A substrate solution is added, which the enzyme converts to a colored product.
- Quantification: The reaction is stopped, and the absorbance is measured. The allergen concentration in the sample is determined by interpolation from the standard curve [1].
DNA-Based Protocol: PCR Detection in Processed Foods
For detecting allergens in processed foods, a robust DNA extraction and amplification protocol is critical. A representative protocol from a study on wheat and maize is as follows [10]:
DNA Extraction:
- Sample Preparation: 100 mg of processed sample (e.g., baked dough ground to a flour-like consistency) is used.
- Lysis: The sample is incubated with CTAB buffer and proteinase K at 65°C.
- Purification: Treatment with RNase A, followed by chloroform extraction.
- Precipitation: DNA is precipitated with a CTAB solution, re-dissolved in NaCl, and re-extracted with chloroform.
- Final Precipitation & Dissolution: DNA is precipitated with isopropanol, washed with 70% ethanol, air-dried, and finally dissolved in 100 μL sterile deionized water.
- Quality Control: DNA concentration and purity are assessed using a UV-Vis spectrophotometer (A260/A280 ratio).
PCR Amplification:
- Primer Design: Primers are designed to target short, stable fragments (~200-300 bp) of the allergen-encoding genes (e.g., HMW-glutenin for wheat, Zea m 14 for maize).
- Reaction Setup: The PCR mixture includes the extracted DNA template, specific primers, dNTPs, and a DNA polymerase.
- Amplification: The reaction is run in a thermal cycler with optimized cycles of denaturation, annealing, and extension.
- Analysis: PCR products are analyzed via agarose gel electrophoresis to confirm the presence and size of the amplicon.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful allergen detection relies on a suite of specialized reagents and tools. The following table details key items used in the featured experiments and their critical functions.
Table 3: Essential Reagents and Materials for Allergen Detection Research
| Reagent / Material | Function | Example Use Case |
|---|---|---|
| CTAB (Cetyltrimethyl ammonium bromide) Buffer | A detergent-based lysis buffer for efficient plant cell wall breakdown and DNA extraction, particularly from complex, polysaccharide-rich matrices. [10] | DNA extraction from wheat and maize flour and baked goods. [10] |
| Bst DNA Polymerase | An enzyme with strong strand displacement activity, essential for isothermal amplification methods like LAMP and LMTIA. It operates at a constant temperature, negating the need for a thermal cycler. [9] | Used in Proofman-LMTIA for rapid, on-site detection of soybean and wheat allergens. [9] |
| Proteinase K | A broad-spectrum serine protease that degrades proteins and inactivates nucleases during DNA extraction, thereby protecting the target DNA from degradation. [10] | Sample incubation step in CTAB-based DNA extraction protocols. [10] |
| Specific Primers & Probes | Short, single-stranded DNA sequences designed to bind complementarily to a unique segment of the target allergen gene, ensuring high detection specificity. [10] [9] | Targeting the Lectin gene for soybean and the GAG56D gene for wheat in duplex Proofman-LMTIA. [9] |
| Hydroxynaphthol Blue (HNB) / Cresol Red (CR) | Metalochromic and pH-sensitive dyes, respectively, used for visual, colorimetric detection of LAMP amplicons without needing to open the reaction tube, reducing contamination risk. [13] | Pre-added to LAMP reactions; a color change (e.g., purple-to-blue for HNB) indicates a positive amplification. [13] |
| Capture & Detection Antibodies (ELISA) | The core components of an ELISA kit. The capture antibody immobilizes the target allergen, while the enzyme-conjugated detection antibody generates a measurable signal, enabling highly specific protein quantification. [1] | Commercial ELISA kits for gluten detection, officially adopted by the Codex Alimentarius. [1] |
The comparative analysis clearly indicates that there is no single superior method for all allergen detection scenarios. The choice between protein-based and DNA-based detection is context-dependent. Protein-based methods like ELISA are indispensable when the goal is direct quantification of the allergenic protein, providing the most biologically relevant data for risk assessment, especially in raw or minimally processed foods. However, for highly processed foods where proteins may be denatured, DNA-based methods (PCR, LAMP, LMTIA) offer a more robust and sensitive alternative due to the greater stability of DNA. Emerging technologies like multiplex allergen microarrays [8] and isothermal amplification with visual detection [13] [9] are pushing the boundaries of multiplexing, speed, and ease of use. The optimal strategy for researchers and industry professionals often involves a complementary use of both techniques, leveraging the strengths of each to ensure the highest level of accuracy and consumer protection in allergen management.
Food allergy has become a significant global public health issue, with increasing prevalence worldwide and no specific treatment available beyond strict avoidance of allergenic foods [1]. This reality places critical importance on accurate food allergen detection for effective regulatory compliance and consumer protection. Within this landscape, two primary analytical approaches have emerged: protein-based methods that directly detect allergenic proteins, and DNA-based methods that take an indirect approach by targeting species-specific genetic markers [1]. While protein-based methods like ELISA (Enzyme-Linked Immunosorbent Assay) detect the actual allergenic molecules, DNA-based techniques identify the genetic blueprint that signals the potential presence of these allergens, making them particularly valuable when proteins have been denatured during food processing [1] [5].
The fundamental distinction between these approaches lies in their analytical targets. Protein-based methods detect the causative agents of allergic reactions but face challenges when protein structures are altered during food processing. In contrast, DNA-based methods exploit the greater stability of DNA molecules under harsh processing conditions, providing an indirect but highly reliable indicator of allergenic ingredient presence [5]. This article provides a comprehensive comparative analysis of DNA-based detection methodologies, their operational principles, and their performance relative to protein-based approaches in food allergen analysis.
Fundamental Principles of DNA-Based Detection
Genetic Markers as Detection Targets
DNA-based allergen detection operates on the principle of identifying unique, species-specific genetic sequences that serve as markers for the presence of allergenic ingredients. Unlike protein-based methods that target the allergenic molecules themselves, DNA-based approaches detect the genetic material from the allergenic source, providing an indirect confirmation of potential allergen presence [1] [14].
The core advantage of this strategy lies in the inherent stability of DNA molecules. DNA typically retains its molecular integrity better than proteins through various food processing treatments, including high-temperature operations [1] [5]. This stability makes DNA-based detection particularly effective for analyzing highly processed foods where protein structures may have been denatured or altered, compromising antibody recognition in immunoassays [1].
Key Genetic Marker Technologies
Several DNA marker technologies have been adapted for food allergen detection, each with distinct characteristics and applications:
PCR-Based Methods: Polymerase Chain Reaction (PCR), particularly quantitative PCR (qPCR), represents the most widely utilized DNA-based approach for allergen detection. These methods amplify and detect specific DNA sequences unique to allergenic sources, providing both qualitative identification and quantitative assessment [1] [5]. qPCR has been adopted as an official analytical tool for food allergen detection in several countries, including Germany and Japan [1].
DNA Biosensors: Emerging DNA-based biosensors incorporate functional DNA strands (such as aptamers), DNA hybridization systems, or DNA templates as recognition elements. These platforms offer advantages including rapid detection, high sensitivity, and potential for on-site analysis when combined with technologies like microfluidics [1] [15] [14].
Isothermal Amplification Methods: Techniques like loop-mediated isothermal amplification (LAMP) and closed-tube saltatory rolling circle amplification provide alternatives to traditional PCR, enabling rapid detection without complex thermal cycling equipment [1]. These methods are particularly suited for point-of-care testing scenarios.
The following workflow illustrates the typical process for DNA-based allergen detection:
Comparative Analysis: DNA-Based vs. Protein-Based Methods
Performance Characteristics Across Methodologies
The selection between DNA-based and protein-based allergen detection methods depends on multiple factors including the food matrix, processing methods, required sensitivity, and regulatory context. The table below summarizes the key characteristics of each approach:
Table 1: Comparison of Food Allergen Detection Methods
| Parameter | DNA-Based Methods (PCR) | Protein-Based Methods (ELISA) | Biosensors (Emerging) |
|---|---|---|---|
| Target | Species-specific DNA sequences | Allergenic protein structures | Varies (DNA, protein, or both) |
| Detection Principle | Nucleic acid amplification and detection | Antigen-antibody interaction | Signal transduction from biorecognition event |
| Sensitivity | High (capable of detecting trace DNA) | High (capable of detecting trace proteins) | Variable (potentially very high) |
| Effect of Food Processing | More stable to thermal processing | Protein denaturation may affect detection | Depends on recognition element |
| Quantification | Indirect correlation with allergen amount | Direct measurement of protein | Variable depending on design |
| Specificity | High species specificity | Potential cross-reactivity with related proteins | Can be highly specific |
| Detection Time | 2-4 hours (including DNA extraction) | 1-2 hours | Minutes to hours |
| Throughput | Moderate to high | High | Potentially high |
| Cost | Moderate | Moderate | Varies widely |
| Regulatory Acceptance | Official method in some countries (e.g., Germany, Japan) | Widely accepted (reference method for some allergens) | Emerging validation |
Detection Limits and Matrix Effects
Comparative studies have demonstrated that the performance of DNA-based methods varies significantly across different food matrices and allergen sources. Research on celery detection using commercial DNA-based test kits revealed that while these kits could detect celery DNA down to 1 ppm spiked protein in five different product groups, a clear matrix effect was observed across different food types [5]. The study evaluated products representing different segments of the AOAC food-matrix triangle, including meat products, snacks, sauces, dried herbs and spices, and smoothies.
Notably, quantification of celery content proved challenging across all food product groups using DNA-based quantification methods, with converted DNA results frequently leading to overestimation of the actual celery amount [5]. This highlights a significant limitation of DNA-based approaches: the variable relationship between DNA content and allergenic protein content in different tissues and processing scenarios.
In comparison studies of detection limits, lateral flow immunoassays (LFI) often showed superior sensitivity for specific allergens, but DNA-based methods like the ATP+ADP+AMP (A3) test provided valuable alternatives for detecting food debris where specific LFIs are not commercially available [16]. The A3 test demonstrated preferable or comparable detection limits to LFIs for crustacean shellfish and for processed grains (with exceptions of wheat flour and buckwheat) [16].
Experimental Protocols in DNA-Based Allergen Detection
Standardized qPCR Protocol for Celery Detection
The following protocol, adapted from comparative kit assessment studies, outlines the standard methodology for DNA-based celery detection [5]:
Sample Preparation and DNA Extraction:
- Homogenize 30 mg of food sample in a 2 mL tube
- Add 1 mL of CTAB buffer, 40 μL of proteinase K (20 mg/mL), and 5 μL of RNase (100 mg/mL)
- Incubate for 90 minutes at 65°C to lyse cells and degrade proteins and RNA
- Centrifuge for 10 minutes at 16,000×g
- Transfer 300 μL of supernatant to a Maxwell cartridge
- Purify DNA using Maxwell RSC 48 instrument with PureFood GMO and authentication method
- Elute DNA in 100 μL elution buffer
- Quantify DNA concentration using spectrophotometry (e.g., Nanodrop 1000)
qPCR Amplification and Detection:
- Prepare reaction mix containing Taqman Universal Master Mix
- Add primers and probe targeting celery-specific markers (e.g., Cel-MDH sequences)
- Use final primer and probe concentrations of 300 nM and 200 nM, respectively
- Add 5 μL of diluted DNA sample (10 ng/μL) to 20 μL reaction mix
- Perform amplification with the following thermal profile:
- Initial steps: 2 minutes at 50°C, 10 minutes at 95°C
- 45 cycles of: 15 seconds at 95°C, 1 minute at 60°C
- Analyze amplification curves and determine Cq values
Data Interpretation:
- Compare Cq values to standard curve from known celery concentrations
- Account for matrix effects through appropriate controls
- Consider potential overestimation when converting DNA results to protein equivalents
Emerging DNA Biosensor Protocol
Advanced DNA-based biosensors are incorporating innovative materials and approaches:
Aptamer-Based Electrochemical Sensor Development:
- Immobilize thiol-modified DNA aptamers on gold electrode surfaces
- Alternatively, adsorb single-stranded DNA aptamers on graphene oxide (GO) surfaces through π-π stacking
- Functionalize aptamers with reporter molecules (methylene blue, ferrocene)
- Upon target binding, monitor conformational changes in aptamers through electrochemical impedance
- Detect signal changes proportional to target concentration [15]
The Scientist's Toolkit: Essential Reagents and Materials
Table 2: Key Research Reagents for DNA-Based Allergen Detection
| Reagent/Material | Function | Examples/Specifications |
|---|---|---|
| CTAB Buffer | Lysis buffer for DNA extraction, helps remove polysaccharides and polyphenols | Contains cetyltrimethylammonium bromide, often with EDTA and sorbitol |
| Proteinase K | Proteolytic enzyme that degrades proteins and nucleases | Typically used at 20 mg/mL concentration |
| RNase A | Ribonuclease that removes contaminating RNA | Typically used at 100 mg/mL concentration |
| DNA Purification Kits | Isolation of high-quality DNA from complex matrices | Maxwell RSC PureFood GMO and Authentication Kit |
| Taq Polymerase | Enzyme for PCR amplification | Thermostable DNA polymerase with buffer system |
| qPCR Master Mix | Optimized mixture for quantitative PCR | Contains Taq polymerase, dNTPs, buffer, often with UDG prevention |
| Sequence-Specific Primers/Probes | Target recognition and amplification | Celery: Cel-MDH-iF/R/probe; Must be validated for specificity |
| DNA Molecular Markers | Size standards for electrophoresis | Typically 50-1000 bp range for fragment analysis |
| Reference Materials | Quality control and quantification | Certified reference materials with known allergen content |
Technological Advances and Future Perspectives
Emerging Trends in DNA-Based Detection
The field of DNA-based allergen detection continues to evolve with several promising technological developments:
Miniaturization and Microfluidics: Integration of DNA-based detection with microfluidic platforms enables assay miniaturization, reducing reagent consumption and analysis time while potentially enabling point-of-care testing [17]. These systems concentrate target molecules into small volumes or regions, enhancing signal strength and improving detection sensitivity [17].
Enhanced Signal Amplification: Novel nucleic acid amplification strategies including hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA) provide enzyme-free signal amplification, improving sensitivity without requiring protein enzymes [15]. These methods enable exponential signal amplification through controlled, autonomous hybridization events.
DNA Nanotechnology: Programmable DNA nanostructures such as DNA tetrahedra and DNA origami provide precise spatial organization for recognition elements, improving binding efficiency and assay performance [15]. These structures offer remarkable addressability and adjustable rigidity for enhanced biosensing applications.
Multiplexing Capabilities: Advanced DNA-based platforms increasingly enable simultaneous detection of multiple allergens in a single reaction, addressing the complex nature of food matrices and potential cross-contamination scenarios [1] [14].
Integration Approaches and Method Selection
The future of food allergen detection likely lies in integrated approaches that leverage the complementary strengths of both DNA-based and protein-based methods. DNA-based techniques excel at specific identification of allergenic sources, particularly in processed foods, while protein-based methods directly quantify the allergenic molecules themselves [1] [3].
The following decision framework illustrates the method selection process:
For researchers and food safety professionals, method selection should consider the specific application context, with DNA-based methods providing critical advantages for specific scenarios but requiring careful interpretation in quantitative applications. As DNA-based technologies continue to advance, particularly in biosensing and miniaturization, their role in comprehensive allergen detection strategies is likely to expand significantly, offering enhanced capabilities for protecting allergic consumers through accurate food labeling and safety assurance.
Current Market Trends and the Dominance of Established Methodologies
The increasing global prevalence of food allergies has positioned food allergen testing as a critical component of modern food safety protocols, with the global market value projected to grow significantly from USD 970.3 million in 2025 to USD 2,062.6 million by 2035 [18]. Within this expanding landscape, a clear dichotomy exists between protein-based and DNA-based detection methodologies. Enzyme-Linked Immunosorbent Assay (ELISA) and Polymerase Chain Reaction (PCR) have emerged as the dominant technologies, accounting for substantial market shares [19] [18]. This guide provides a comparative analysis of these established methodologies, examining their technical principles, performance under experimental conditions, and respective roles in current market trends. Designed for researchers and drug development professionals, it offers a detailed examination of the protocols and reagent solutions that underpin reliable allergen detection.
The food allergen testing market is characterized by steady growth, driven by rising allergy prevalence, stringent regulatory requirements, and increasing consumer demand for safe, accurately labeled food products [20] [18]. Within this framework, established methodologies maintain their dominance through proven reliability and continuous innovation.
Market Size and Growth Trajectory
- United States Market: Projected to reach US$ 451.58 million by 2033, growing at a CAGR of 7.00% from 2024 [20].
- Global Market: Expected to expand from USD 970.3 million in 2025 to USD 2,062.6 million by 2035, reflecting a CAGR of 7.8% [18].
- Technology Leadership: PCR-based testing leads the technology segment with a 35.4% market share in 2025, while immunoassay-based methods like ELISA remain the gold standard for routine screening [18] [4].
Dominant Testing Methodologies
The market is segmented by technology, with key methods including:
- PCR (Polymerase Chain Reaction): Valued for its high specificity in detecting allergen-specific DNA sequences.
- ELISA (Enzyme-Linked Immunosorbent Assay): Recognized as the gold standard for protein detection, known for high sensitivity and specificity.
- Lateral Flow Devices (LFD): Used for rapid, on-site screening.
- Mass Spectrometry (e.g., LC-MS/MS): Gaining traction for its high precision in quantifying specific allergenic proteins [6] [3].
Table 1: Market Position of Major Allergen Testing Technologies
| Technology | Market Share (2025) | Primary Detection Target | Key Strength |
|---|---|---|---|
| PCR | 35.4% [18] | DNA (Allergen-specific genes) | High specificity, effective for processed foods [19] [7] |
| ELISA/Immunoassays | Dominant position [4] | Proteins (Allergenic proteins) | High sensitivity and specificity, cost-effective [19] [4] |
| Lateral Flow Devices | Reported | Proteins | Rapid, on-site results |
| Mass Spectrometry | Emerging | Proteins (Specific protein markers) | High precision and multiplexing capability [6] |
Comparative Methodological Analysis
A nuanced understanding of the performance characteristics of protein-based (e.g., ELISA) and DNA-based (e.g., PCR) methods is essential for selecting the appropriate analytical tool for a given food matrix and regulatory purpose.
Performance Characteristics and Experimental Data
Direct comparative studies and market analyses reveal distinct performance profiles for each methodology.
Table 2: Experimental Performance Comparison of ELISA vs. PCR
| Performance Parameter | ELISA (Protein-Based) | PCR (DNA-Based) |
|---|---|---|
| Detection Limit | Parts per million (ppm) levels [4] | As low as 0.01 ng/mL for some targets [6] |
| Effect of Food Processing | Proteins can be denatured, leading to potential false negatives [4] | DNA is more stable; effective in processed foods (e.g., baked at 220°C) [7] |
| Specificity | High, but can cross-react with related proteins [4] | Very high, based on specific DNA sequences [7] |
| Quantification | Direct quantification of protein levels | Quantifies DNA, which is a proxy for allergen presence |
| Time to Result | Relatively fast (hours) [20] | Can be slower due to DNA extraction and amplification (hours to days) [20] |
| Cost-Effectiveness | More economical for routine screening [4] | Higher cost due to equipment and consumables [19] [20] |
Detailed Experimental Protocol: PCR Detection of Allergens
A 2025 study on detecting wheat and maize allergens provides a robust experimental model for DNA-based method validation [7].
Experimental Workflow
The following diagram illustrates the key steps in the PCR-based detection of allergens in processed foods.
- Sample Preparation: Wheat and maize dough samples were baked at 180°C and 220°C for 10 to 60 minutes to simulate processing.
- DNA Extraction: The CTAB (cetyltrimethyl ammonium bromide) method was used to isolate genomic DNA from 100 mg of each sample. DNA concentration and purity were assessed via spectrophotometry (NanoDrop One).
- PCR Amplification: Novel primers were designed to target major allergen genes:
- Wheat: High-molecular-weight (HMW) and low-molecular-weight (LMW) glutenin subunits.
- Maize: Zea m 14, Zea m 8, and zein.
- Analysis: PCR products were analyzed using agarose gel electrophoresis.
- Key Findings:
- DNA degradation correlated with increased baking temperature and time.
- The optimal primers successfully detected allergen genes even after baking at 220°C for 40-60 minutes.
- PCR sensitivity was positively influenced by DNA extractability and gene copy number but decreased with longer amplicon length and more intense processing.
The Scientist's Toolkit: Key Research Reagent Solutions
Successful experimentation in allergen detection relies on a suite of specialized reagents and instruments.
Table 3: Essential Research Reagents and Materials for Allergen Testing
| Reagent/Material | Function/Application | Example in Context |
|---|---|---|
| CTAB Buffer | DNA extraction from complex food matrices; helps in separating DNA from polysaccharides and proteins. | Used in the protocol for isolating DNA from baked wheat/maize dough [7]. |
| Proteinase K & RNase A | Enzymes that degrade contaminating proteins and RNA during DNA extraction, purifying the target DNA. | Part of the CTAB-based DNA extraction protocol to obtain high-quality genomic DNA [7]. |
| ELISA Kits | Ready-to-use kits containing antibodies specific to allergenic proteins (e.g., casein, Ara h 2) for quantitative detection. | Used for routine screening of allergens like gluten, milk, egg, and peanut in various food products [4]. |
| PCR Primers | Short, single-stranded DNA sequences designed to bind to and amplify specific allergen genes. | Primers for wheat HMW-glutenin and maize zein genes were critical for specific detection [7]. |
| Taq DNA Polymerase | Enzyme that synthesizes new DNA strands during PCR, essential for amplifying the target DNA sequence. | A core component of any PCR master mix for allergen gene amplification. |
| Antibodies (for ELISA) | Proteins that bind specifically to target allergenic proteins, enabling their detection and quantification. | Monoclonal antibodies are used in kits like the SENSIStrip Gluten PowerLine to reduce false negatives [20]. |
Underlying Mechanism of IgE-Mediated Food Allergy
Understanding the biological mechanism that allergen detection methods are designed to guard against is crucial for contextualizing their application. The following pathway delineates the immunologic process of an IgE-mediated allergic reaction, which is the primary mechanism for the food allergies detected by these methodologies [2].
Pathway Explanation: The process begins with Allergen Exposure and initial Sensitization Phase, where the allergen is presented by Antigen-Presenting Cells (APCs), leading to T-helper 2 (Th2) cell activation. This prompts B cells to produce allergen-specific Immunoglobulin E (IgE) antibodies, which then bind to Fcε receptors on mast cells and basophils [2]. Upon Re-exposure to the same allergen, the Effector Phase is triggered: the allergen cross-links the IgE antibodies on the mast cell surface, causing degranulation and the release of inflammatory mediators like histamine and leukotrienes, which ultimately produce the clinical symptoms of an allergic reaction [2].
Discussion and Future Outlook
The comparative analysis confirms that the dominance of established methodologies like ELISA and PCR is not a matter of stagnation but a reflection of their complementary and validated roles in a complex market. ELISA remains the uncontested leader for routine, high-throughput protein detection due to its cost-effectiveness and direct measurement of the allergenic moiety [4]. In contrast, PCR provides an indispensable tool for scenarios where protein-based methods falter, such as in highly processed foods where proteins are denatured but DNA remains amplifiable [7] [4].
The future evolution of this field will be shaped by several key trends. Multiplexed assays that can detect multiple allergens simultaneously are gaining prominence, improving efficiency and reducing costs [6] [18]. Furthermore, the integration of artificial intelligence (AI) and machine learning is poised to enhance data analysis, improve prediction models, and potentially reduce interpretation errors [19] [6]. There is also a growing demand for portable, rapid on-site testing devices that provide results in real-time, empowering manufacturers with immediate quality control data [18]. Finally, advanced proteomic techniques like mass spectrometry are establishing a new benchmark for precision, capable of simultaneously quantifying specific allergenic proteins with high accuracy, thus complementing the existing methodological toolkit [6] [3].
In conclusion, the current market trends do not indicate a wholesale replacement of one established methodology by another, but rather a strategic coexistence and integration. The choice between protein-based and DNA-based methods is, and will remain, contingent on the specific analytical question, the nature of the food matrix, regulatory requirements, and the necessary balance between cost, speed, and accuracy.
Methodologies in Practice: From ELISA and LC-MS/MS to PCR and Isothermal Amplification
Food allergies represent a significant and growing global public health concern, compelling the need for reliable detection methods to ensure food safety and comply with labelling regulations [3]. The accurate detection of food allergens relies on two primary analytical approaches: protein-based methods, which directly target the allergenic proteins themselves, and DNA-based methods, which detect allergen-specific DNA sequences as an indirect marker [1]. Within the sphere of protein-based detection, immunoassays are paramount due to their specificity, sensitivity, and adaptability. This guide provides a comparative analysis of three key immunoassay formats—the Enzyme-Linked Immunosorbent Assay (ELISA), Lateral Flow Devices (LFDs), and emerging Immunosensors—framed within the broader context of protein-based versus DNA-based detection strategies. Aimed at researchers and scientists, this article offers an objective comparison of performance characteristics, detailed experimental protocols, and essential reagent toolkits.
Performance Comparison at a Glance
The following tables summarize the core characteristics and quantitative performance data of the discussed methods, allowing for a quick, objective comparison.
Table 1: Overall Method Comparison for Allergen Detection
| Feature | ELISA | Lateral Flow Devices (LFDs) | Immunosensors | DNA-based Methods (qPCR) |
|---|---|---|---|---|
| Detection Principle | Immunological, colorimetric | Immunological, visual/reader | Electrochemical, optical | Nucleic acid amplification |
| Detection Target | Proteins | Proteins | Proteins | DNA |
| Throughput | High | Low to Moderate | Moderate | High |
| Assay Time | Several hours | 10-30 minutes | Minutes to an hour | Several hours |
| Ease of Use | Requires training | Simple, minimal training | Varies; can be complex | Requires specialized training |
| Quantification | Excellent | Semi-quantitative / Qualitative | Excellent | Quantitative (indirect) |
| Portability | No | Yes | Yes (many formats) | No |
| Best Use Case | Regulatory compliance, high-sensitivity quantitation | Rapid screening, point-of-use testing | Rapid, sensitive quantitation, multiplexing | Detection of thermally processed allergens where proteins may degrade [1] |
Table 2: Experimental Performance Data from Comparative Studies
| Method / Assay Type | Target | Reported Sensitivity | Reported Specificity | Key Findings / Context |
|---|---|---|---|---|
| ELISA (IgG) | SARS-CoV-2 Antibody | 81.5% | 100% | Used as a benchmark in a clinical study [21]. |
| ELISA (IgA) | SARS-CoV-2 Antibody | 93.1% | 80.6% | Demonstrated higher sensitivity but lower specificity than IgG ELISA in the same study [21]. |
| Lateral Flow (LFI) | SARS-CoV-2 Antibody | Variable: 61.2% - 91.8% | Variable: 61.7% - 80.2% | Performance is highly dependent on the commercial brand and design [21] [22]. |
| Electrochemiluminescence (CLIA) | SARS-CoV-2 Antibody | 91.8% | 80.2% | Showed superior sensitivity compared to ELISA and LFDs in a head-to-head study [22]. |
| DNA-based Kits (qPCR) | Celery Allergen | Detects down to 1 ppm spiked protein | High, but affected by matrix | Successful detection, but quantitative performance was challenging across different food matrices [5]. |
Detailed Methodologies and Experimental Protocols
Protein-Based Detection: Immunoassays
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA is a foundational, plate-based immunological technique renowned for its high sensitivity and robust quantification capabilities. The following outlines a standard sandwich ELISA protocol, commonly used for allergen detection [1].
Protocol: Sandwich ELISA for Allergen Detection
- Plate Coating: A microplate is coated with a capture antibody specific to the target allergen. The plate is then incubated overnight, typically at 4°C, to allow passive adsorption.
- Blocking: After washing to remove unbound antibody, the plate is blocked with a protein-based solution (e.g., Bovine Serum Albumin or casein) to cover any remaining protein-binding sites on the plastic surface, preventing non-specific binding in later steps.
- Sample Incubation: The food sample extract, appropriately diluted in buffer, is added to the well. If the target allergen is present, it binds to the immobilized capture antibody. The plate is incubated (e.g., 1-2 hours at 37°C) and then washed.
- Detection Antibody Incubation: A second enzyme-conjugated detection antibody, specific to a different epitope on the same allergen, is added. This forms an antibody-allergen-antibody "sandwich." Another incubation and wash step follows.
- Signal Development: A colorless enzyme substrate is added. The conjugated enzyme (e.g., Horseradish Peroxidase) converts the substrate into a colored product.
- Signal Measurement & Quantification: The enzymatic reaction is stopped, and the color intensity (Absorbance) is measured with a plate reader. The concentration of the allergen in the sample is determined by interpolation from a standard curve run concurrently.
The workflow of this sandwich ELISA procedure is systematized in the diagram below.
Lateral Flow Devices (LFDs)
Lateral Flow Devices are rapid, single-use tests designed for qualitative or semi-quantitative analysis. Their operation is based on the capillary flow of a liquid sample across a strip containing immobilized reagents [23].
Protocol: Lateral Flow Immunoassay Operation
- Sample Application: The liquid sample (e.g., food extract) is applied to the sample pad. The pad may contain buffers to adjust the sample pH and ensure optimal flow and binding conditions.
- Conjugate Release: As the sample migrates, it rehydrates and mobilizes conjugated detector particles (e.g., colloidal gold or colored latex beads) stored in the conjugate pad. These conjugates are antibodies specific to the target allergen, bound to the reporter particle.
- Formation of Complexes: If the target allergen is present in the sample, it binds to the mobilized conjugated antibodies, forming an analyte-detector complex.
- Capture at Test Line: The complex continues to flow along the nitrocellulose membrane until it reaches the test line. This line is striped with a second, immobilized capture antibody specific to the target allergen. The complex is trapped, accumulating the colored particles and producing a visible line.
- Capture at Control Line: The remaining fluid and any excess conjugate continue to the control line, which contains an antibody that captures the detector particles regardless of the presence of the allergen (e.g., an anti-species antibody). This line must always appear to validate the test.
- Result Interpretation: The appearance of both the control and test lines indicates a positive result. Only the control line indicates a negative result. If the control line fails to appear, the test is invalid.
The components and process flow of a typical sandwich-format LFD are illustrated below.
Immunosensors
Immunosensors are analytical devices that integrate an immunological recognition element (antibody) with a physicochemical transducer to produce a digital electronic signal. Biosensors are regarded as a promising detection method due to their rapidity, high sensitivity, and potential for on-site use [1].
Protocol: Electrochemical Immunosensor Operation
While designs vary, a general protocol for an electrochemical immunosensor is as follows:
- Bio-recognition Layer Preparation: The surface of the electrode is modified with a capture antibody specific to the target allergen. This can be achieved through various chemical immobilization techniques to ensure stable and oriented antibody attachment.
- Blocking: The electrode surface is blocked with a non-reactive protein (e.g., BSA) to minimize non-specific adsorption of other proteins from the sample matrix.
- Sample Incubation: The prepared food extract is applied to the sensor surface. The target allergen, if present, binds to the capture antibodies. The sensor is then washed.
- Signal Transduction: The binding event is measured by the transducer. In a common electrochemical format, an enzyme-labeled secondary antibody is added. Upon adding an electrochemical substrate, the enzyme catalyzes a reaction that produces an electrical current (amperometry) or changes the electrical potential/impedance (potentiometry/electrochemical impedance spectroscopy). The magnitude of this change is proportional to the allergen concentration.
- Data Analysis: The signal is processed by associated electronics and software, providing a quantitative readout of the allergen concentration.
DNA-Based Allergen Detection
For comparison, DNA-based methods like quantitative Polymerase Chain Reaction (qPCR) provide an indirect approach to detecting the presence of an allergenic ingredient.
Protocol: qPCR for Allergen Detection (e.g., Celery)
- DNA Extraction: DNA is isolated from the food matrix using a commercial kit, such as the Maxwell RSC PureFood GMO and Authentication Kit, which may involve CTAB buffer, proteinase K, and RNase incubation [5].
- DNA Quantification and Quality Assessment: The concentration and purity of the extracted DNA are determined using a spectrophotometer (e.g., Nanodrop) [5].
- qPCR Setup: The reaction mix is prepared containing a DNA polymerase (e.g., Taqman Universal Master Mix), sequence-specific primers, and a fluorescently-labeled probe (e.g., FAM-BBQ) designed to bind a unique gene sequence of the allergenic source (e.g., celery) [5].
- Amplification and Detection: The plate is run in a real-time PCR instrument. The thermal cycling process amplifies the target DNA. The probe is cleaved during amplification, releasing a fluorescent signal monitored in real-time.
- Quantification: The cycle threshold (Ct), at which the fluorescence crosses a background threshold, is determined. The Ct value is inversely proportional to the amount of target DNA in the sample. Quantification is achieved by comparison to a standard curve from known concentrations of the target DNA [5].
The Scientist's Toolkit: Key Research Reagent Solutions
Successful development and execution of these assays depend on critical reagents. The table below details essential materials and their functions.
Table 3: Essential Reagents for Immunoassay Development
| Reagent / Material | Function | Key Considerations |
|---|---|---|
| Capture & Detection Antibodies | Specific recognition and binding of the target analyte (allergen). | Affinity & Specificity are paramount to avoid false positives/negatives [23]. Clonality: Monoclonal antibodies offer consistency; Polyclonal antibodies can increase sensitivity through multi-epitope recognition [23]. Supply: Must be scalable for commercial production [23]. |
| Reporter Molecules | Generate a detectable signal indicating analyte presence. | Colloidal Gold: Intense color, easy conjugation, visual readout [23]. Latex Beads: Can be colored or fluorescent, allow for multiplexing [23]. Enzymes (e.g., HRP): Used in ELISA for signal amplification via substrate conversion [1]. Fluorescent Dyes: Enable high-sensitivity quantification, require readers [23]. |
| Microplates & Membranes | Solid support for assay construction. | ELISA Plates: High protein-binding capacity polystyrene. LFD Membranes: Nitrocellulose is standard; its flow rate and protein-binding capacity are critical [23]. |
| Sample Pads & Conjugate Pads | LFD components for sample application and reagent release. | Sample Pad: Must efficiently absorb and filter the sample matrix [23]. Conjugate Pad: Must stably store and efficiently release the labeled antibody conjugate [23]. |
| Blocking Agents | Prevent non-specific binding to surfaces. | Proteins like Bovine Serum Albumin (BSA), casein, or synthetic polymers are used to coat unused binding sites on plates and membranes [23]. |
| Control Reagents | Validate the correct performance of the test. | Typically, an antibody that binds the labeled detector antibody irrespective of the analyte (e.g., an anti-species antibody) is striped as a control line [23]. |
The choice between ELISA, LFDs, and immunosensors for food allergen detection is dictated by the specific application requirements. ELISA remains the gold standard for high-sensitivity, quantitative analysis in a laboratory setting, while LFDs offer unparalleled speed and simplicity for rapid screening. Immunosensors represent the cutting edge, promising to combine the sensitivity of laboratory methods with the speed and portability of point-of-use tests. When contextualized within the protein-based vs. DNA-based debate, immunoassays provide a direct measure of the allergenic protein, making them highly relevant for risk assessment. However, as with any method, careful validation for each specific food matrix is essential to ensure accurate and reliable results [21] [5]. The ongoing innovation in reagents, materials, and detection technologies will continue to enhance the performance and accessibility of these critical food safety tools.
Food allergies are a significant and growing public health concern worldwide, affecting an estimated 1-8% of the population depending on geographic region and age group [3]. For allergic individuals, strict avoidance of allergenic foods remains the primary management strategy, making accurate food labeling and detection of unintended cross-contamination critical for consumer safety [24] [25]. This comparative analysis focuses on two principal technological approaches for allergen detection in food products: protein-based methods (specifically targeted mass spectrometry) and DNA-based methods (primarily PCR-based techniques). The fundamental distinction between these approaches lies in their analytical targets: mass spectrometry (MS) directly detects and quantifies the allergenic proteins themselves, while DNA-based methods detect genetic markers associated with allergenic foods [26] [1] [27].
The "Big 8" allergens—comprising milk, eggs, fish, crustacean shellfish, tree nuts, peanuts, wheat, and soybeans—are responsible for the majority of significant allergic reactions and are subject to mandatory labeling in many countries [24] [25]. Accurately quantifying these allergens in complex food matrices presents substantial analytical challenges, particularly at the low concentrations (parts-per-million to parts-per-billion) that can still trigger reactions in sensitive individuals. This guide provides a comprehensive comparison of targeted proteomics and DNA-based methods, examining their respective technical capabilities, limitations, and appropriate applications within food safety testing protocols.
Fundamental Principles and Methodological Comparison
Protein-Based Detection: Targeted Mass Spectrometry
Targeted proteomics using mass spectrometry, specifically Multiple Reaction Monitoring (MRM) also known as Selected Reaction Monitoring (SRM), represents a sophisticated approach for the direct quantification of allergenic proteins [24] [25]. This methodology employs triple quadrupole mass spectrometers, where the first and third quadrupoles act as mass filters for specific precursor and product ions, respectively, while the second quadrupole functions as a collision cell [25]. The core principle involves quantifying proteotypic peptides—unique peptide sequences that reliably indicate the presence of a specific allergenic protein—through their mass-to-charge ratio (m/z) and fragmentation patterns [25].
The critical innovation in quantitative targeted proteomics is the use of stable isotope-labeled internal standards (AQUA peptides), which are chemically identical to the target peptides but differ in mass [24]. These standards are added at known concentrations during sample preparation, enabling precise absolute quantification of the target allergenic proteins by comparing the mass spectrometric response of the native peptide to its labeled counterpart. This approach allows for multiplexed allergen quantification, where multiple allergenic proteins from different food sources can be detected and quantified simultaneously in a single analysis [24] [6].
DNA-Based Detection: PCR Methods
DNA-based detection methods, primarily polymerase chain reaction (PCR) and its variants (e.g., real-time PCR), represent an indirect approach to allergen detection that targets species-specific DNA sequences rather than the allergenic proteins themselves [26] [1] [27]. The fundamental principle involves extracting DNA from food samples, amplifying target sequences using sequence-specific primers, and detecting the amplified products. In real-time PCR, the accumulation of amplified DNA is monitored throughout the reaction using fluorescent dyes or probes, allowing for both detection and quantification of the target DNA [1].
While DNA-based methods do not directly measure the allergenic proteins, they provide an effective surrogate marker for the presence of allergenic ingredients, particularly in complex matrices where protein integrity may be compromised [26]. The technique capitalizes on the greater stability of DNA compared to proteins, especially in processed foods where proteins may be denatured or structurally altered, potentially affecting antibody recognition in immunoassays [1] [27].
Comparative Workflow Analysis
The following diagram illustrates the core procedural differences between mass spectrometry-based and DNA-based allergen detection workflows:
Experimental Protocols and Methodologies
Targeted Mass Spectrometry Workflow for Allergen Quantification
Sample Preparation and Protein Extraction
The initial step involves extracting proteins from the food matrix using appropriate extraction buffers. The composition of these buffers is critical and often needs to be optimized for different food matrices (e.g., baked goods, dairy, meat products) to ensure efficient protein recovery while minimizing interference from carbohydrates, lipids, and other food components [24] [25]. For complex matrices, additional cleanup steps such as precipitation or immunoaffinity depletion may be employed to reduce background interference and enhance sensitivity [25].
Protein Digestion and Peptide Standardization
Extracted proteins are denatured, reduced, and alkylated before enzymatic digestion. Trypsin is the most commonly used protease due to its high specificity for cleaving C-terminal to lysine and arginine residues, generating peptides of optimal length for LC-MS analysis [25]. Stable isotope-labeled internal standard peptides (AQUA peptides) are added at this stage at known concentrations, which correct for variations in sample preparation and ionization efficiency during MS analysis [24].
Liquid Chromatography and Mass Spectrometric Analysis
The complex peptide mixture is separated by reversed-phase liquid chromatography, typically using nano-flow or capillary LC systems to enhance sensitivity [24] [25]. Eluting peptides are ionized via electrospray ionization (ESI) and introduced into the mass spectrometer. In the MRM/SRM workflow, the triple quadrupole mass spectrometer is programmed to monitor specific precursor ion → product ion transitions for each target peptide, typically 3-5 transitions per peptide [25]. The mass filters are set to these predefined m/z values, and detection occurs only during retention time windows specific to each peptide, significantly enhancing sensitivity and specificity compared to full-scan methods [25].
Data Analysis and Quantification
Peak areas for the target peptides and their corresponding isotope-labeled standards are integrated using specialized software. Protein quantification is achieved by comparing the peak area ratio of the native peptide to the internal standard against a calibration curve generated from standards of known concentration [24] [25]. Data analysis includes verification of transition peak area ratios and co-elution to ensure proper peptide identification and minimize false positives [25].
DNA-Based Detection Workflow
DNA Extraction and Purification
DNA is extracted from food samples using commercial kits or established protocols, with careful consideration to break down the food matrix and remove inhibitors that could affect downstream PCR amplification [1] [27]. The quality and quantity of extracted DNA are assessed, and additional purification steps may be required for complex matrices.
PCR Amplification and Detection
Species-specific primers are designed to target conserved or unique genomic sequences of the allergenic food source [1]. In real-time PCR, amplification is monitored using intercalating dyes (e.g., SYBR Green) or sequence-specific fluorescent probes (e.g., TaqMan). The cycle threshold (Ct) value, representing the PCR cycle at which fluorescence exceeds background levels, is used for quantification relative to standard curves [1].
Performance Comparison and Experimental Data
Quantitative Comparison of Detection Methods
Table 1: Comparative Performance Characteristics of Allergen Detection Methods
| Parameter | Targeted Mass Spectrometry | DNA-Based Methods (PCR) | ELISA (Reference) |
|---|---|---|---|
| Detection Limit | 0.1-5 mg/kg (ppm) [25] | Varies by target; generally similar sensitivity [1] | 0.1-5 mg/kg (ppm) [25] |
| Target Analyte | Proteins/peptides (direct detection of allergens) [25] | DNA (indirect marker) [26] [1] | Proteins (antibody recognition) [1] [25] |
| Multiplexing Capacity | High (dozens of allergens simultaneously) [24] [6] | Moderate (typically single or few targets per reaction) [1] | Low (typically single allergen per test) [25] |
| Impact of Food Processing | Moderate (proteotypic peptides often survive processing) [25] | Low (DNA more stable than proteins) [1] [27] | High (epitopes may be denatured) [25] |
| Quantification Approach | Absolute quantification with isotope standards [24] | Relative quantification to standard curves [1] | Interpolation from standard curves [25] |
| Specificity Challenges | Minimal with proper peptide selection [25] | Cross-reactivity with related species [1] | Antibody cross-reactivity [25] |
Applicability Across Food Allergen Types
Table 2: Method Performance Across Major Allergen Categories
| Allergen Category | Targeted MS Applications | DNA-Based Detection Applications | Notable Considerations |
|---|---|---|---|
| Peanut | Quantification of Ara h 1, Ara h 2, Ara h 3 [25] | Detection of peanut-specific DNA sequences [1] | MS provides protein-specific profile; DNA confirms peanut presence |
| Milk | Independent quantification of casein and whey proteins [25] | Detection of bovine DNA [1] | MS can distinguish different milk protein fractions |
| Egg | Quantification of Gal d 1 (ovalbumin) and Gal d 2 (ovomucoid) [6] | Detection of chicken DNA [1] | Egg-specific proteins vs. general poultry detection |
| Seafood | Detection of parvalbumin (fish) and tropomyosin (shellfish) [27] | Species-specific fish/shellfish DNA detection [27] | MS targets conserved allergen proteins; DNA identifies species |
| Tree Nuts | Detection of specific allergenic proteins in walnut, hazelnut, etc. [25] | Detection of species-specific DNA [1] | Complex matrix effects for both methods |
The Researcher's Toolkit: Essential Reagents and Materials
Core Reagents and Instrumentation
Table 3: Essential Research Tools for Allergen Detection Methods
| Category | Specific Reagents/Equipment | Function/Purpose |
|---|---|---|
| Mass Spectrometry | Triple quadrupole mass spectrometer (QQQ) | MRM/SRM analysis for targeted quantification [25] |
| Nano/Capillary LC system | High-sensitivity peptide separation [24] | |
| Stable isotope-labeled peptides (AQUA) | Internal standards for absolute quantification [24] | |
| Trypsin (sequencing grade) | Protein digestion to generate measurable peptides [25] | |
| DNA-Based Methods | Real-time PCR instrument | DNA amplification and detection [1] |
| Species-specific primers and probes | Target amplification with specificity [1] | |
| DNA extraction/purification kits | Isolation of amplifiable DNA from food matrices [1] | |
| Inhibition-resistant polymerase | Reliable amplification from complex food matrices [1] | |
| Data Analysis | BatMass [28] | Mass spectrometry data visualization |
| vMS-Share [29] | Public MS repository and data mining | |
| Allergen Peptide Browser [25] | Resource for proteotypic peptide selection |
Critical Analysis and Future Perspectives
Targeted mass spectrometry offers significant advantages for allergen detection, particularly its ability to directly quantify specific allergenic proteins with high specificity and multiplexing capability [24] [25]. The technology demonstrates robustness across various food matrices and can be more resistant to the effects of food processing compared to antibody-based methods, as it targets peptide sequences rather than conformational epitopes [25]. Furthermore, MS methods can be adapted to monitor specific protein modifications (oxidation, deamidation, glycation) that may occur during processing, providing insights into how these modifications affect allergenicity [25].
DNA-based methods excel in scenarios where protein integrity is compromised but DNA remains detectable, particularly in highly processed foods [1] [27]. They also offer advantages in cost and accessibility for routine testing of single allergens. However, they cannot directly quantify the specific proteins responsible for allergic reactions and may detect DNA from allergenic sources that don't contain problematic levels of proteins [26].
The emerging field of biosensors and rapid detection technologies shows promise for future allergen detection, potentially combining the specificity of immunological recognition with the sensitivity of advanced signal detection systems [6] [1]. Additionally, computational approaches and bioinformatics resources such as the Allergen Peptide Browser are enhancing the selection of optimal proteotypic peptides for MS assays, facilitating method development and standardization across laboratories [25].
In conclusion, both targeted mass spectrometry and DNA-based methods provide valuable capabilities for allergen detection, with their respective advantages making them complementary rather than competing technologies. The selection of an appropriate method depends on the specific analytical requirements, including the need for direct protein quantification, the complexity of the food matrix, the degree of food processing, and the required throughput and multiplexing capability. As both technologies continue to evolve, they will collectively enhance our ability to protect consumers through accurate allergen detection and quantification.
Within food safety and public health, accurate food allergen detection is paramount for protecting sensitive individuals. The detection methodologies primarily fall into two categories: protein-based methods (e.g., ELISA, mass spectrometry), which detect the allergenic proteins themselves, and DNA-based methods, which detect the DNA sequences encoding these proteins [1] [3]. DNA-based detection, primarily using Polymerase Chain Reaction (PCR) techniques, offers a powerful alternative, particularly for processed foods where proteins may be denatured but DNA remains amplifiable [1] [7]. This guide provides a comparative analysis of three foundational PCR techniques—Endpoint, Real-Time, and Multiplex PCR—within the context of food allergen detection research. It objectively compares their performance, supported by experimental data, and details protocols relevant to scientists and drug development professionals engaged in food safety analysis.
Core Principles and Comparative Analysis of PCR Techniques
The fundamental principle of PCR is to enzymatically amplify a specific DNA target, making it detectable and quantifiable. The techniques discussed here apply this principle in different ways, each with distinct advantages for allergen detection.
Endpoint PCR: This is the traditional form of PCR. After amplifying the target DNA over a set number of cycles, the total accumulated product is analyzed via agarose gel electrophoresis. Measurement occurs in the plateau phase of the PCR reaction, where reagents have become depleted and the amplification rate is no longer consistent [30]. This makes it inherently less suitable for reliable quantification.
Real-Time PCR (qPCR): Also known as quantitative PCR (qPCR), this method monitors the amplification of DNA in real-time as the reaction occurs. Fluorescent dyes or probes report the amount of PCR product after each cycle. By collecting data during the exponential phase of amplification, where the reaction is most consistent and reproducible, qPCR allows for precise quantification of the initial DNA template [31] [30]. The key output is the Cycle threshold (Ct), the cycle number at which the fluorescence crosses a predefined threshold, which is inversely correlated to the starting amount of target DNA [32].
Multiplex PCR: This variant allows for the simultaneous amplification of multiple different DNA targets in a single reaction tube by incorporating multiple pairs of specific primers [33]. This is highly valuable for efficiently screening a food product for several potential allergens at once, saving time, reagents, and sample material.
Table 1: Comparative Performance of Endpoint, Real-Time, and Multiplex PCR for Food Allergen Detection.
| Feature | Endpoint PCR | Real-Time PCR (qPCR) | Multiplex PCR |
|---|---|---|---|
| Quantification Capability | Semi-quantitative or qualitative | Fully quantitative | Typically qualitative or semi-quantitative |
| Sensitivity | Lower | 100-fold more sensitive than endpoint PCR [31] | High, comparable to endpoint or qPCR depending on detection method |
| Dynamic Range | ~2 orders of magnitude [31] | >4 orders of magnitude [31] | Dependent on the number of targets and detection system |
| Precision (CV) | ~6.8% (fluorescence intensity) [31] | ~0.14% (Ct value) [31] | Varies with optimization |
| Key Advantage | Simplicity, low cost | Accuracy, wide dynamic range, high throughput | Multi-target analysis, high efficiency |
| Primary Limitation | Low resolution, low throughput | Higher cost, complex data analysis | Requires extensive optimization, primer competition [33] |
| Best for Allergen Detection | Qualitative presence/absence checks | Precise quantification and high-sensitivity detection [1] | Screening for multiple allergens simultaneously [33] |
Experimental Data in Food Allergen Detection
Sensitivity and Specificity in Practice
The superior sensitivity of real-time PCR is a critical factor in allergen detection, where trace amounts can trigger reactions. A comparative study demonstrated that a real-time PCR system was 100-fold more sensitive than an end-point PCR system when using a single mouse liver cDNA standard [31]. Furthermore, the precision of real-time PCR, with a percentage standard error of the mean of 0.14% for the Ct value across 30 replicates, far exceeded the 6.8% observed for end-point fluorescence intensity [31]. This high precision is crucial for generating reliable and reproducible data in quantitative food safety assessments.
The extreme sensitivity of qPCR, however, requires careful interpretation in a diagnostic context. A clinical study on Pneumocystis pneumonia found that real-time PCR had a significantly higher false-positive rate (35%) compared to endpoint PCR (10%), likely due to its enhanced ability to detect low-level colonization that does not constitute active disease [34]. This underscores the importance of interpreting qPCR results for allergens within the context of known thresholds for eliciting an allergic reaction [3].
Impact of Food Processing on Detection
Food processing, which can degrade DNA, presents a challenge for PCR-based detection. Research on detecting wheat and maize allergens has shown that the length of the DNA amplicon is a critical factor. Successful detection in baked goods is associated with shorter amplicons. For instance, in maize samples baked at 220°C for 40 minutes, specific primers for allergens like Zea m 14 and zein were able to generate positive PCR results, whereas longer genomic DNA fragments were degraded [7]. This confirms the established best practice of designing assays with short amplicons (~200–300 bp) for the analysis of processed foods [7].
Table 2: Experimental Data from PCR-based Allergen Detection Studies.
| Study Focus | PCR Method | Key Experimental Finding | Implication for Allergen Detection |
|---|---|---|---|
| Fundamental Sensitivity [31] | Endpoint vs. Real-Time | Real-time PCR was 100x more sensitive. Dynamic range: >4 logs (qPCR) vs. ~2 logs (Endpoint). | qPCR is superior for detecting trace amounts of allergenic ingredients. |
| Diagnostic Specificity [34] | Real-Time PCR | False-positive rate of 35% vs. 10% for endpoint PCR in a clinical pathogen model. | High sensitivity may detect irrelevant contamination; results require clinical/contextual correlation. |
| Processed Food Analysis [7] | Endpoint PCR | Reliable detection of wheat/maize allergens in samples baked at 220°C using short, specific amplicons. | Assay design (amplicon size) is critical for detecting allergens in processed foods. |
| Multi-Species Screening [33] | Multiplex PCR | Simultaneous detection of 6 meat species (beef, chicken, pork, etc.) in raw and processed food products. | Efficient for screening food products for multiple potential allergenic contaminants. |
Detailed Experimental Protocols
Protocol for Relative Quantification Using Real-Time PCR (qPCR)
Relative quantification is a common approach in gene expression and allergen detection, which compares the amount of target allergen DNA to a reference gene DNA [35] [32].
- Sample Preparation and DNA Extraction: Extract genomic DNA from the food sample using a validated method, such as CTAB-based extraction for complex matrices [7]. Assess DNA concentration and purity using spectrophotometry.
- Assay Design: Design and validate primer sets (and probes if using a probe-based system) for both the target allergen gene (e.g., a glutenin gene for wheat) and a stable reference gene (e.g., a plant housekeeping gene). Amplicons should be short (100-200 bp) for robustness [7].
- PCR Efficiency Calculation: Prepare a serial dilution of a known positive DNA sample. Run the qPCR assay for these dilutions in triplicate. Plot the average Ct values against the logarithm of the dilution factor. The PCR efficiency (%) is calculated from the slope of the standard curve: Efficiency = (10^(-1/slope) - 1) × 100 [32]. An efficiency between 90-110% is typically acceptable.
- Experimental Run: Perform qPCR on test samples, including no-template controls. Each sample should be run with both the target and reference gene assays.
- Data Analysis (Livak Method): This common method assumes the PCR efficiencies of the target and reference genes are approximately equal and close to 100% [32].
- Calculate ∆Ct for each sample: ∆Ct = Ct(target) - Ct(reference)
- Calculate ∆∆Ct: ∆∆Ct = ∆Ct(treated sample) - ∆Ct(control calibrator sample)
- Calculate the expression ratio (fold change): Ratio = 2^(-∆∆Ct)
Protocol for Developing a Multiplex PCR Assay for Allergen Screening
Multiplex PCR requires careful optimization to amplify several targets simultaneously without artifacts [33].
- Primer Design: Design primer pairs for each allergen target (e.g., peanut, hazelnut, sesame). Ensure all primers have similar melting temperatures (Tm) and do not interact with each other (to avoid primer-dimers). Design amplicons to be of distinct lengths for clear separation by gel electrophoresis.
- Initial Optimization of Singleplex Reactions: Optimize each primer pair individually in a singleplex PCR reaction to determine its ideal annealing temperature and MgCl2 concentration.
- Multiplex Reaction Assembly: Combine all primer pairs in a single tube. The concentration of each primer may need to be adjusted relative to singleplex conditions to balance the amplification efficiency of all targets.
- Thermal Cycling Optimization: Perform a gradient PCR to find the annealing temperature that allows all primers to bind specifically to their respective targets. The number of cycles may be increased compared to a singleplex assay.
- Analysis: Analyze the PCR products by capillary electrophoresis or agarose gel electrophoresis. The presence of distinct bands of the expected sizes confirms the detection of specific allergens.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Key Research Reagent Solutions for PCR-Based Allergen Detection.
| Reagent/Material | Function | Example in Application |
|---|---|---|
| Sequence-Specific Primers | To define the target DNA region for amplification. | Primers for wheat HMW-glutenin or maize Zea m 14 genes [7]. |
| DNA Polymerase (Taq) | Enzyme that synthesizes new DNA strands. | Thermostable polymerase for robust PCR cycling. |
| dNTPs | Building blocks (nucleotides) for new DNA strands. | Included in the master mix for DNA synthesis. |
| Fluorescent Probes/Dyes | For real-time detection of amplified DNA (e.g., SYBR Green, TaqMan). | SYBR Green dye binding to double-stranded DNA amplicons [35]. |
| Buffer Components (MgCl₂) | Provides optimal ionic environment and co-factors for polymerase activity. | MgCl₂ concentration is critical for multiplex PCR optimization [33]. |
| Internal Amplification Control (IAC) | Non-target DNA sequence to confirm the PCR reaction was not inhibited. | Added to the reaction to rule to false negatives [33]. |
| CTAB Lysis Buffer | For efficient DNA extraction from complex, polysaccharide-rich food matrices. | Used to extract DNA from wheat and maize flour [7]. |
Endpoint, Real-Time, and Multiplex PCR each occupy a distinct and valuable niche in the DNA-based detection of food allergens. Endpoint PCR remains a cost-effective tool for simple, qualitative checks. Real-Time PCR is the undisputed choice for sensitive, precise quantification, essential for benchmarking against legal thresholds, though its results must be interpreted wisely. Multiplex PCR offers unparalleled efficiency for comprehensive screening programs. The choice of technique is not a matter of which is universally "best," but which is most fit-for-purpose, depending on the required level of quantification, the number of allergens to be monitored, the nature of the food matrix, and the available resources. Within the broader context of food allergen research, these DNA-based methods provide a robust, complementary approach to protein-based detection, enhancing the reliability of food safety controls and the protection of public health.
The accurate detection of food allergens is a critical challenge in ensuring food safety and public health. Traditional detection paradigms have long relied on protein-based methods, such as enzyme-linked immunosorbent assay (ELISA), which target the allergenic proteins themselves. In contrast, DNA-based methods detect allergen-specific genetic sequences, offering a fundamentally different approach. Within this context, emerging nucleic acid amplification techniques (NAATs), particularly isothermal methods such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA), represent a significant advancement. Their integration with microfluidic technology has created powerful, portable diagnostic platforms that are reshaping the landscape of food allergen testing [36] [37] [6].
This guide provides a comparative analysis of LAMP and RPA technologies, focusing on their operational principles, performance metrics, and practical applications when integrated into microfluidic systems. It is designed to equip researchers and drug development professionals with the experimental data and protocols necessary to evaluate these DNA-based methods against traditional protein-based detection for allergen analysis.
LAMP and RPA are two prominent isothermal amplification techniques that eliminate the need for thermal cycling, a major limitation of conventional PCR. This makes them exceptionally suitable for point-of-care testing (POCT) and resource-limited settings [38] [39].
Loop-Mediated Isothermal Amplification (LAMP): Developed by Notomi et al. in 2000, LAMP uses a DNA polymerase with high strand displacement activity and 4 to 6 primers that recognize 6 to 8 distinct regions of the target DNA. The reaction occurs at a constant temperature of 60–65°C and can produce up to 10^9–10^10 copies of the target in under an hour. The complex primer design contributes to high specificity, but the technique can be susceptible to non-specific amplification and primer-dimer formation, which may lead to false positives [38] [40] [41].
Recombinase Polymerase Amplification (RPA): Commercialized in 2006, RPA operates at a much lower temperature range of 37–42°C. It employs recombinase enzymes that facilitate primer insertion into homologous DNA sequences, followed by strand-displacement synthesis. A key advantage is its simplicity, requiring only two primers, and its rapid amplification, often completing in 20 minutes or less. However, it is also prone to primer-dimer artifacts and non-specific amplification, especially in complex samples [42] [38] [40].
Table 1: Fundamental Characteristics of LAMP and RPA
| Feature | LAMP | RPA |
|---|---|---|
| Year Developed | 2000 [38] | 2006 [38] |
| Reaction Temperature | 60–65°C [38] [41] | 37–42°C [42] [40] |
| Typical Assay Time | 30–60 minutes [38] | 20–40 minutes [43] [40] |
| Number of Primers | 4–6 [38] [41] | 2 [42] |
| Key Enzyme | Bst DNA Polymerase [38] | Recombinase (e.g., T4 UvsX) [38] |
| Main Advantage | High amplification efficiency, robustness | Low temperature, speed |
| Main Challenge | Complex primer design, false positives [40] [41] | Non-specific amplification, primer-dimer formation [42] [40] |
Performance Comparison and Experimental Data
When deployed for pathogen detection—a proxy for the rigor required in allergen analysis—LAMP and RPA demonstrate high sensitivity and specificity. Their performance is further enhanced when coupled with CRISPR-based systems for detection.
Sensitivity and Specificity
A 2025 study on Group B Streptococcus (GBS) detection directly compared LAMP- and RPA-based CRISPR/Cas12b assays. The findings indicated that the LAMP-CRISPR/Cas12b system consistently outperformed the RPA-based system across all template concentrations, particularly at low copy numbers (30 and 10 copies per test) [42]. To address the sensitivity limitations of RPA, the researchers developed a one-pot, two-temperature protocol (RPA at 39°C followed by Cas12b activation at 62°C), which significantly improved its low-copy detection rate, achieving a sensitivity of 10 copies per test [42]. Clinical validation with 60 samples showed excellent concordance with standard methods (96.7% vs. culture; 98.3% vs. qPCR) [42].
In seafood authentication, a real-time microfluidic RPA chip was developed to identify Atlantic cod, sablefish, and toothfish. This platform demonstrated high sensitivity, detecting genomic DNA at concentrations as low as 10^3 fg/μL and recombinant plasmid at 10 copies/μL. When applied to 141 commercial products, it achieved 100% identification accuracy for the target species [44].
Addressing False Positives
A significant challenge for both techniques is distinguishing true positives from non-specific amplification (NSA). High-Resolution Melt (HRM) analysis post-amplification has been successfully used to differentiate specific and non-specific products for both LAMP and RPA [40]. This closed-tube, post-amplification method relies on the sequence-dependent melt temperature (Tm) of amplicons, where shorter NSA products (e.g., from primer dimers) melt at lower temperatures (~50-60°C) than longer, specific targets (70-90°C) [40].
Table 2: Comparative Performance Data of LAMP and RPA Platforms
| Application Target | Technology | Sensitivity | Specificity/Accuracy | Key Experimental Finding |
|---|---|---|---|---|
| Group B Streptococcus [42] | LAMP-CRISPR/Cas12b | Outperformed RPA at low copies (10 copies/test) | 98.3% concordance with qPCR | A two-temperature protocol (39°C → 62°C) improved RPA performance. |
| Influenza Viruses [41] | RT-LAMP on Microfluidic Chip | 10^2 copies/μL | High consistency with RT-qPCR on 296 clinical samples | The closed-chip system prevented aerosol contamination and false positives. |
| Seafood Authentication [44] | Real-time RPA on Microfluidic Chip | 10^3 fg/μL (gDNA); 10 copies/μL (plasmid) | 100% identification accuracy | Successfully identified mislabeling in 31.91% of 141 commercial samples. |
| General Pathogen Detection [40] | LAMP/RPA with HRM | Effective in stochastic regime (0.1 ng total DNA) | Able to distinguish specific from non-specific amplification | HRM analysis was completed in under 4 minutes on a microfluidic instrument. |
Microfluidic Integration and Experimental Workflows
Microfluidic technology automates and miniaturizes complex laboratory processes onto a single chip, making it an ideal platform for integrating LAMP and RPA into portable, user-friendly systems [43] [39].
Microfluidic Chip Architectures
Two primary configurations are used:
- Centrifugal Microfluidic Chips: These chips use rotational force to move liquids through microchannels. They are highly integrated and suitable for high-throughput parallel processing [44] [41]. For example, a 4-channel LAMP chip can initiate reagent mixing and reaction upon centrifugation, allowing simultaneous detection of multiple targets [41].
- Digital Microfluidics (DMF): Based on the electrowetting-on-dielectric (EWOD) principle, DMF manipulates discrete droplets on an array of electrodes. This "lab-on-a-chip" approach offers flexible, programmable control over multiple steps like sample preparation, amplification, and detection in a single, compact system with minimal human intervention [43].
Representative Experimental Protocols
This protocol is for multiplex influenza virus detection and can be adapted for allergen gene targets.
- Primer Design: Design inner (FIP/BIP), outer (F3/B3), and loop (LF/LB) primers targeting conserved regions of the allergen gene using software like Primer Explorer V5.
- Chip Priming: Pre-load the LAMP reaction mix (containing Bst polymerase, dNTPs, buffer, and intercalating dye) and specific primer sets into the respective channels of the polycarbonate microfluidic chip in a contamination-free environment.
- Sample Loading: Introduce the extracted DNA or RNA sample (for RT-LAMP) into the chip's sample inlet.
- Centrifugal Activation: Place the chip in the instrument, which heats the base to 65°C. The chip is then spun at a low speed (e.g., 1600 rpm) to mix the liquids and eliminate bubbles, followed by high-speed spins (e.g., 4500 rpm) to centrifuge the mixture into the final reaction chambers.
- Amplification & Detection: Incubate the chip at 65°C for 40 minutes. Fluorescence is monitored in real-time. A positive signal indicates the presence of the target allergen gene.
This protocol describes an extraction-free, one-pot assay suitable for simple and rapid testing.
- Sample Preparation: Subject the sample (e.g., swab eluent) to a simple pretreatment like heat lysis (95°C for 5 min) to release DNA without formal extraction.
- Reaction Setup: Prepare a single reaction tube containing all RPA reagents (recombinase, polymerase, primers) and CRISPR/Cas12b reagents (AapCas12b enzyme, crRNA, fluorescent reporter).
- Amplification Phase: Incubate the reaction tube at 39°C for 40 minutes for the RPA amplification to occur.
- CRISPR Detection Phase: Increase the temperature to 62°C for 5 minutes to activate the trans-cleavage activity of Cas12b. If the target DNA is present, the Cas12b-crRNA complex binds to it and cleaves the fluorescent reporter.
- Signal Readout: Visualize the fluorescence signal under UV or blue light. A positive reaction can be seen with the naked eye.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of these techniques requires a specific set of reagents and materials. The table below lists key components for setting up LAMP and RPA experiments.
Table 3: Essential Research Reagents and Materials for LAMP and RPA
| Item Name | Function/Description | Example Application |
|---|---|---|
| Bst DNA Polymerase | The core enzyme for LAMP, possessing strand displacement activity. | Catalyzing DNA amplification at a constant 60-65°C [38] [41]. |
| RPA Basic Kit | Contains recombinase (e.g., T4 UvsX), single-stranded DNA-binding protein, and strand-displacing polymerase. | Enabling rapid isothermal amplification at 37-42°C [42] [38]. |
| AapCas12b Enzyme | A thermostable Cas protein used for specific nucleic acid detection after amplification. | Generating a fluorescent signal via trans-cleavage in a one-pot assay [42]. |
| Fluorescent DNA Intercalator (e.g., SYBR Green) | A dye that fluoresces upon binding to double-stranded DNA. | Real-time monitoring of LAMP or RPA amplification [40] [41]. |
| exo Probe (for RPA) | A specific probe cleaved by exonuclease III during RPA, generating a target-specific fluorescence signal. | Enabling real-time fluorescent detection of RPA products [44]. |
| Microfluidic Chip (Polycarbonate) | A disposable device with micro-channels and chambers that automate fluid handling and reactions. | Creating a closed, integrated system for sample-to-answer detection [41]. |
| crRNA (CRISPR RNA) | A short RNA guide that directs the Cas protein to a specific DNA target sequence. | Providing sequence-specificity in the CRISPR-based detection step [42]. |
LAMP and RPA, particularly when enhanced by CRISPR systems and integrated into microfluidic platforms, represent a new generation of DNA-based detection tools. While LAMP generally offers higher amplification efficiency and robustness, RPA excels in speed and lower operational temperature. The choice between them depends on the specific application requirements, including available infrastructure, required throughput, and the complexity of the sample matrix. For food allergen testing, these DNA-based methods provide a compelling alternative to traditional protein-based assays, offering high sensitivity, specificity, and the potential for rapid, on-site deployment to ensure food safety and regulatory compliance.
Navigating Analytical Challenges: Matrix Effects, Processing, and Cross-Reactivity
Impact of Food Processing on Allergen Protein Integrity and DNA Amplification
Food allergies represent a significant and growing global public health concern, affecting individuals of all ages and requiring strict avoidance of allergenic foods for sensitive individuals [1] [3]. Accurate detection of food allergens is therefore crucial for consumer protection, regulatory compliance, and food labeling. Detection methodologies primarily fall into two categories: protein-based methods that directly target allergenic proteins and DNA-based methods that detect allergen-encoding genes [1] [7]. However, food processing techniques—including thermal treatment, high-pressure processing, and fermentation—profoundly impact the integrity and detectability of both proteins and DNA [3] [7]. These processing-induced alterations present substantial challenges for allergen detection, influencing the reliability, sensitivity, and applicability of different analytical approaches.
This comparative guide provides an objective analysis of protein-based versus DNA-based detection methods, focusing specifically on how food processing affects allergen protein integrity and DNA amplifiability. We present experimental data comparing method performance under various processing conditions, detail standardized protocols for evaluating processing impacts, and provide evidence-based recommendations for method selection in different food matrices. Understanding these factors is essential for researchers, food safety specialists, and regulatory professionals to make informed decisions about allergen detection strategies for both routine analysis and method development.
Comparative Analysis of Detection Methods
Fundamental Principles and Technical Characteristics
Protein-based methods directly target the causative agents of allergic reactions—the allergenic proteins themselves. These techniques include enzyme-linked immunosorbent assays (ELISA), lateral flow immunoassays (LFI), Western blotting, and mass spectrometry (LC-MS/MS) [1] [3]. ELISA operates on the principle of antibody-antigen recognition, providing quantitative results with high sensitivity and specificity for intact proteins [1]. LFI offers rapid, on-site screening based on the same immunological principles but is typically qualitative or semi-quantitative [16]. Mass spectrometry provides highly specific identification and quantification of allergen peptides, often with the ability to detect multiple allergens simultaneously [1].
DNA-based methods, primarily polymerase chain reaction (PCR) and its quantitative variant (qPCR), indirectly detect allergens by targeting species-specific DNA sequences [1] [5]. These methods amplify and detect conserved genes or allergen-encoding DNA sequences, providing excellent specificity for the biological source of the allergen [7]. PCR is particularly valuable for detecting allergens from closely related species when specific antibodies are unavailable [5]. The fundamental difference in targets—proteins versus DNA—underlies the divergent performance characteristics of these methods, especially in processed foods where the stability of these molecules differs significantly.
Table 1: Core Characteristics of Protein-based and DNA-based Allergen Detection Methods
| Characteristic | Protein-based Methods (ELISA) | DNA-based Methods (PCR) |
|---|---|---|
| Detection Target | Intact protein structures (epitopes) | Species-specific DNA sequences |
| Primary Applications | Quantification of allergenic protein; Regulatory compliance | Species identification; Detection when proteins are denatured |
| Typical Sensitivity | ppm to ppb levels for most allergens [1] | Varies; can detect <10 target copies [5] |
| Specificity | Epitope-dependent; cross-reactivity possible [1] | Species-dependent; can distinguish closely related species [5] |
| Quantification Capability | Direct (standard curves for proteins) | Indirect (correlation to DNA copy number) |
| Throughput | Moderate to high | Moderate |
| Cost per Analysis | Moderate to high | Low to moderate |
Impact of Processing on Target Molecules
Food processing induces structural changes that differentially affect proteins and DNA, directly influencing detection efficacy. Thermal processing denatures proteins, altering their three-dimensional conformation and potentially destroying antibody-binding epitopes essential for immunological detection [3]. For example, baking at 180-220°C causes progressive degradation of both genomic DNA and target allergen genes, with detectability decreasing as temperature and duration increase [7]. However, DNA demonstrates greater stability than proteins under certain thermal processing conditions, maintaining its molecular integrity when proteins may become undetectable [1] [5].
Chemical processing (e.g., fermentation, acidic/alkaline treatments) can hydrolyze proteins into peptides too small for antibody recognition, leading to false-negative results in immunoassays [3]. DNA is generally more resistant to such chemical degradation, though extreme pH conditions can cause fragmentation [7]. High-pressure processing and irradiation can modify protein structures while potentially preserving DNA sequences suitable for amplification [3].
The following experimental workflow illustrates a standardized approach for evaluating processing impacts on both detection targets:
Performance Comparison in Processed Foods
Direct comparison studies demonstrate that processing conditions significantly influence the relative performance of protein versus DNA-based detection methods. Research on wheat and maize allergens revealed that while baking at 220°C for 60 minutes degraded genomic DNA, target allergen genes (HMW-GS and LMW-GS for wheat; Zea m 14, Zea m 8, and zein for maize) remained detectable with appropriately designed PCR protocols [7]. The same thermal processing would likely denature many protein epitopes, reducing immunoassay sensitivity.
A comparative assessment of celery detection in complex food matrices found that DNA-based methods could detect celery at 1 ppm spiked protein levels across five different product categories, though matrix effects significantly influenced detection capability [5]. Notably, protein-based ELISA methods for celery are often unavailable due to cross-reactivity with other Apiaceae family species like carrot and parsley [5]. This highlights a key advantage of DNA-based methods: their ability to discriminate between closely related species when protein cross-reactivity poses challenges for immunological approaches.
For soy allergen detection in complex foods, a novel electrochemical sensor using molecularly imprinted polymers demonstrated accurate detection across 42 different food products representing over 300 ingredients, correctly identifying soy presence regardless of processing methods including heat, fermentation, and acidity [45]. This emerging technology shows particular promise for detecting allergens in processed matrices where conventional methods may fail.
Table 2: Processing Impact on Detection Method Performance
| Processing Method | Impact on Proteins | Impact on DNA | Recommended Detection Approach |
|---|---|---|---|
| Thermal Processing (Baking, Canning) | Denaturation, epitope destruction [3] | Fragmentation with prolonged exposure [7] | DNA-based for intense processing; Protein-based for mild processing [1] |
| Fermentation | Hydrolysis, structural modification [3] | Generally stable; possible nuclease activity | DNA-based methods typically preferred [1] |
| High-Pressure Processing | Conformational changes, potential epitope alteration [3] | Minimal impact; strand breaks possible | Both methods viable; protein-based if epitopes preserved |
| Chemical Processing (Acid/Alkaline) | Denaturation, hydrolysis [3] | Acid-induced depurination possible | DNA-based methods generally more reliable |
| Extrusion | Severe denaturation, aggregation | Significant fragmentation | DNA-based with short amplicon targets (<200 bp) [7] |
Experimental Data and Validation
Thermal Processing Effects on DNA Detectability
Controlled studies investigating wheat and maize allergen detection demonstrate clear relationships between processing intensity and DNA amplification efficiency. Research examining baking effects at 180°C and 220°C over 60 minutes established that DNA detectability decreases with increasing temperature and processing duration [7]. However, appropriate primer selection targeting shorter amplicons (typically <200-300 bp) maintained detection sensitivity even after extensive thermal processing [7].
In these experiments, wheat HMW-GS and LMW-GS genes remained detectable after 60 minutes of baking at 220°C when using optimized primers. Similarly, maize allergen genes (Zea m 14, Zea m 8, and zein) were detectable after 40 minutes of baking at 220°C [7]. The experimental protocol involved:
- Sample Preparation: Dough samples prepared from wheat and maize flour, baked at controlled temperatures (180°C and 220°C) for timed intervals (10-60 minutes)
- DNA Extraction: CTAB-based method with proteinase K and RNase treatment, chloroform extraction, and isopropanol precipitation
- PCR Amplification: Species-specific primers for allergen genes, with amplicon size optimized for processed samples
- Detection: Agarose gel electrophoresis and spectrophotometric quantification
This systematic approach demonstrates that despite DNA degradation during processing, target genes remain amplifiable when analytical parameters are optimized for the specific processing conditions [7].
Method Comparison in Complex Food Matrices
Evaluations of detection methods in multi-ingredient food products reveal significant matrix effects influencing both protein and DNA-based approaches. A comprehensive assessment of three commercial DNA-based test kits for celery detection in five food matrix categories ((plant-based) meat products, snacks, sauces, dried herbs and spices, and smoothies) demonstrated variable performance across matrices despite all kits meeting their specifications [5].
Notably, while all kits detected celery at 1 ppm spiked protein levels, quantification proved challenging across all food product groups, with DNA results tending to overestimate actual celery content [5]. This highlights a critical limitation of DNA-based methods: the indirect relationship between DNA quantity and allergenic protein content, which can vary by tissue type, growth conditions, and processing history.
For soy allergen detection, an electrochemical sensor utilizing molecularly imprinted polymers (MIPs) correctly identified soy presence across 42 different food products with varying complexity, demonstrating consistent performance where traditional ELISA might struggle with processed ingredients [45]. The sensor detected genistein, a soy allergen tracer, at clinically relevant levels (signal-to-noise ratio >1.3) regardless of food matrix composition or processing method.
Decision Framework for Method Selection
The following decision pathway provides a systematic approach for selecting the appropriate detection method based on processing conditions and analytical requirements:
Research Reagent Solutions
Selecting appropriate reagents and materials is crucial for reliable allergen detection in processed foods. The following table details essential research reagents and their applications in evaluating processing impacts on allergen detection.
Table 3: Essential Research Reagents for Allergen Detection Studies
| Reagent/Material | Function/Application | Considerations for Processed Foods |
|---|---|---|
| CTAB Extraction Buffer | DNA isolation from complex matrices [7] | Effective for processed samples; enhances DNA recovery from lipid-rich materials |
| Proteinase K | Protein degradation during DNA extraction [5] [7] | Essential for breaking down processed food matrices and releasing DNA |
| Specific Primers (e.g., Cel-MDH) | Target amplification in qPCR [5] | Should be designed for short amplicons (<200 bp) for processed samples [7] |
| TaqMan Probes | Quantitative PCR detection [5] | Provide specific detection in complex matrices; require sequence knowledge |
| Monoclonal/Polyclonal Antibodies | Protein detection in immunoassays [1] | Epitope recognition must be validated for processed forms of allergens |
| Molecularly Imprinted Polymers | Synthetic recognition elements [45] | Potential resistance to processing-induced changes compared to antibodies |
| Reference Materials | Method calibration and validation [5] | Should represent processed forms when analyzing processed foods |
The comparative analysis of protein-based and DNA-based allergen detection methods reveals a complex interplay between food processing technologies and analytical performance. Protein-based methods provide direct measurement of allergenic proteins but are susceptible to processing-induced epitope destruction, potentially leading to false-negative results in extensively processed foods. DNA-based methods offer advantages for species identification and detection in processed matrices where proteins may be denatured, but they provide only indirect measurement of allergen content and are influenced by DNA fragmentation patterns.
The experimental data presented demonstrates that method selection must be guided by processing conditions, target analytes, and matrix characteristics. For mildly processed foods, protein-based methods typically offer direct and relevant quantification of allergens. For extensively processed products, DNA-based methods with optimized primer design often provide more reliable detection. Emerging technologies like biosensors and molecularly imprinted polymers show promise for bridging this gap, potentially offering robust detection across processing conditions.
Future method development should focus on integrated approaches that combine the strengths of both protein and DNA-based detection, particularly for complex and extensively processed food products. Additionally, standardized reference materials representing processed forms of allergens would significantly improve method validation and comparability across studies. Understanding these fundamental relationships between food processing and allergen detection targets enables researchers and food safety professionals to select optimal strategies for accurate allergen detection in increasingly complex food products.
Overcoming Matrix Interference in Complex and Processed Foods
The accurate detection of food allergens is a critical public health imperative, with an estimated 2-5% of adults and 4-10% of children worldwide affected by food allergies [46]. This diagnostic challenge is particularly pronounced in complex and processed foods, where the food matrix—the physical structure and molecular interactions within a food—can significantly interfere with analytical results [47] [48]. Matrix effects alter how allergens are released during extraction, modified during processing, and detected by analytical methods, potentially leading to both false-positive and false-negative results that directly impact consumer safety [8].
The core of this challenge lies in the fundamental transformation that foods undergo during processing. Techniques such as thermal treatment, high-pressure processing, fermentation, and enzymatic modification can induce profound changes in allergenic proteins, including denaturation, aggregation, Maillard reactions, and epitope masking [8] [3]. These modifications affect protein solubility, antibody recognition patterns, and DNA extractability, creating analytical hurdles that differ significantly between protein-based and DNA-based detection methodologies [49]. Consequently, understanding and overcoming matrix interference has become a central focus in food allergen analysis, driving methodological innovations and comparative assessments of existing technologies.
Analytical Methodologies: Principles and Mechanisms
Protein-Based Detection Systems
Protein-based detection methods target the immunologically active components of food allergens—the specific proteins that trigger IgE-mediated reactions in sensitized individuals [46]. Enzyme-Linked Immunosorbent Assay (ELISA) operates on the principle of antigen-antibody interaction, where antibodies specifically bind to allergenic proteins extracted from food samples [49]. This interaction is visualized through an enzyme-mediated colorimetric reaction, with signal intensity correlating to allergen concentration [49]. ELISA's effectiveness, however, depends heavily on the availability and solubility of intact protein epitopes, which can be compromised by processing-induced structural changes [8].
Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) has emerged as a powerful alternative for direct allergen protein detection [50]. This technique identifies unique peptide markers derived from allergenic proteins through enzymatic digestion, followed by separation and detection based on mass-to-charge ratios [50]. A significant advantage of LC-MS/MS lies in its ability to differentiate between highly similar allergens, such as pistachio and cashew, which often cross-react in immunoassays due to protein homology [50]. Furthermore, multiplex allergen microarray-based immunoassays enable simultaneous detection of multiple allergenic proteins using minimal sample volume, providing a comprehensive profile of IgE-reactive components in complex matrices [8].
DNA-Based Detection Systems
DNA-based methods, primarily Polymerase Chain Reaction (PCR), take an indirect approach to allergen detection by targeting species-specific DNA sequences rather than the allergenic proteins themselves [49] [50]. Through thermal cycling, PCR exponentially amplifies target DNA sequences, enabling detection even in heavily processed foods where proteins may be denatured beyond antibody recognition [49]. The technique is particularly valuable for analyzing highly processed, hydrolyzed, or fermented products where DNA stability exceeds that of proteins [49]. Real-time PCR formats provide both qualitative and semi-quantitative data, though they do not directly measure the immunologically relevant proteins responsible for allergic reactions [50].
Lateral Flow Assays (LFA) offer rapid, on-site screening through immunochromatography, where allergen-antibody complexes form visible lines on a test strip [49]. While technically protein-based, LFAs are often used alongside PCR in complementary testing schemes, providing quick preliminary results that may be confirmed with more specific methods [49].
Table 1: Core Principles of Major Allergen Detection Methods
| Method | Target Analyte | Detection Principle | Key Advantage |
|---|---|---|---|
| ELISA | Specific allergenic proteins | Antigen-antibody interaction with enzyme-mediated signal generation | High sensitivity and specificity for intact proteins |
| LC-MS/MS | Signature peptides from allergenic proteins | Mass spectrometric detection of protein-specific peptides | Direct protein detection unaffected by antibody cross-reactivity |
| PCR | Species-specific DNA sequences | Amplification of target DNA segments | Effective for processed foods where proteins are denatured |
| Lateral Flow Assays | Specific allergenic proteins | Immunochromatographic visualization on membrane strips | Rapid, on-site testing without specialized equipment |
Comparative Performance in Complex Matrices
Methodological Strengths and Limitations
The performance divergence between protein-based and DNA-based methods becomes most apparent when analyzing processed foods with complex matrices. Thermal processing presents a particular challenge for immunoassays, as heat-induced structural changes can mask or destroy conformational epitopes targeted by antibodies, leading to underestimation of allergen content [8]. However, DNA-based methods may also struggle with highly processed matrices where DNA becomes fragmented or bound to other food components, reducing amplification efficiency [50].
Cross-reactivity represents another significant challenge, particularly for antibody-based methods. Research demonstrates that ELISA methods often cannot distinguish between botanically related species such as pistachio and cashew due to protein homology, potentially resulting in false positives [50]. In contrast, LC-MS/MS can differentiate these allergens by targeting unique peptide markers, though method development requires extensive characterization of specific allergenic proteins [50]. PCR assays designed with species-specific primers can also achieve high discrimination between related species, provided sufficient genetic divergence exists in the targeted sequences [50].
The food matrix composition itself introduces substantial interference through biochemical interactions between allergens and other food components. Proteins may become trapped within carbohydrate networks, complex with polyphenols, or undergo Maillard reactions with reducing sugars, all of which affect extractability and detection [8] [48]. Dairy products exemplify this challenge, where the dairy matrix—the complex physical structure of milk, cheese, and yogurt—significantly influences protein bioavailability and detectability [47] [48]. The fat content, protein interactions, and mineral composition in dairy matrices create very different detection environments compared to baked goods or chocolate, where high sugar and fat content can interfere with both protein and DNA extraction [50].
Quantitative Performance Data
Recent validation studies provide compelling quantitative data on method performance across different food matrices. One investigation comparing detection methods for pistachio allergens in various processed foods demonstrated significant performance variations [50]. The LC-MS/MS method achieved a screening detection limit (SDL) of 1 mg/kg across multiple matrices, including chocolate, cereals, and sauces, demonstrating robust performance despite matrix complexity [50].
Table 2: Performance Comparison Across Food Matrices (Pistachio Allergen Detection)
| Food Matrix | ELISA Recovery (%) | PCR Results | LC-MS/MS (SDL: 1 mg/kg) | Key Matrix Interferences |
|---|---|---|---|---|
| Chocolate | 45-60% | False negatives in highly processed samples | Reliable detection | High fat content impedes protein/DNA extraction |
| Breakfast Cereals | 55-70% | Variable detection based on thermal processing | Reliable detection | Maillard reaction products mask protein epitopes |
| Sauces | 70-85% | Consistent detection | Reliable detection | Emulsion structure traps allergens |
| Baked Goods | 40-65% | DNA degradation in crust regions | Reliable detection with optimized extraction | Protein aggregation during baking |
| Meat Products | 60-75% | Consistent detection | Reliable detection | High protein background creates competition |
The data reveal that ELISA recovery rates vary substantially across matrices, with particularly low recovery in high-fat and high-heat processed products [50]. PCR demonstrates more consistent detection in non-thermally processed matrices but shows vulnerability to DNA degradation in extensively heated products like baked goods [50]. LC-MS/MS emerges as the most robust technology across diverse matrices, though it requires sophisticated instrumentation and expert operation [50].
Experimental Workflows for Matrix Challenge Investigation
Method Comparison Protocol
A standardized experimental approach for evaluating matrix interference involves parallel analysis of identical processed food samples using protein-based and DNA-based methods. The following protocol has been validated for assessing allergen detection in complex matrices:
Sample Preparation: Create incurred samples by introducing known quantities of target allergens (e.g., pistachio, milk, egg) into various food matrices (chocolate, baked goods, sauces) prior to processing. Subject samples to different processing conditions (thermal treatment, high-pressure, fermentation) to simulate commercial food production [8] [3].
Protein Extraction: Employ multiple extraction buffers (high-salt, high-pH, commercial extraction solutions) to evaluate the impact of extraction efficiency on method performance. Research indicates that high-salt buffers recover up to 30% more protein from certain plant matrices compared to low-pH buffers [8]. Include a digestion step with trypsin or other proteases for LC-MS/MS analysis to generate target peptides [50].
Parallel Analysis: Divide each extracted sample for simultaneous analysis by ELISA, LC-MS/MS, and PCR. For ELISA, use commercial kits with antibodies against major allergens (e.g., Ara h 1 for peanut, casein for milk) [49]. For LC-MS/MS, target specific proteotypic peptides that uniquely identify each allergen (e.g., Pis v 3.0101 for pistachio) [50]. For PCR, amplify species-specific DNA sequences with demonstrated specificity [49].
Data Analysis: Calculate recovery rates for each method-matrix combination and statistically compare results to identify significant matrix effects. Include positive and negative controls in each batch to ensure method validity [50].
Workflow Visualization
The following diagram illustrates the experimental workflow for evaluating matrix effects in allergen detection:
Experimental Workflow for Matrix Effect Evaluation
Essential Research Reagents and Materials
Successful investigation of matrix interference requires carefully selected reagents and reference materials. The following table details essential components for method comparison studies:
Table 3: Essential Research Reagents for Allergen Detection Studies
| Reagent Category | Specific Examples | Research Function | Considerations for Matrix Studies |
|---|---|---|---|
| Reference Materials | Certified allergenic food powders (peanut, milk, egg), Incurred food reference materials | Method calibration, Quality control | Ensure materials represent processing conditions being studied |
| Extraction Buffers | High-salt buffers (PBS), Urea/thiourea solutions, Commercial extraction cocktails | Protein solubilization, Allergen recovery | Buffer composition significantly impacts extraction efficiency from different matrices |
| Antibodies | Monoclonal antibodies against major allergens (Ara h 1, casein, β-lactoglobulin), Secondary enzyme-conjugated antibodies | ELISA and LFA detection, Epitope recognition | Evaluate multiple antibodies for each allergen to assess epitope stability |
| Proteases | Trypsin, Chymotrypsin, Pepsin | Protein digestion for MS analysis, Simulating gastrointestinal conditions | Digestion efficiency varies with matrix complexity |
| PCR Reagents | Species-specific primers/probes, DNA polymerase, dNTPs, DNA extraction kits | DNA amplification, Species identification | Primers must target multi-copy genes for sensitivity in complex matrices |
| LC-MS Standards | Stable isotope-labeled peptide standards, Mobile phase additives | Mass spectrometry quantification, Chromatographic separation | Labeled standards correct for matrix-induced ionization suppression |
Emerging Technologies and Future Directions
The evolving landscape of allergen detection includes several promising technologies designed to overcome matrix challenges. Multiplexed immunoassays and mass spectrometry are achieving detection limits as low as 0.01 ng/mL for specific allergenic proteins, providing unprecedented sensitivity for trace allergen detection [6]. These advances are particularly valuable for analyzing products with complex ingredient profiles where multiple allergens may be present simultaneously.
Artificial intelligence-enhanced platforms are being developed to predict allergenicity of novel ingredients and optimize detection strategies based on matrix composition [6]. Non-destructive techniques such as hyperspectral imaging (HSI) and Fourier Transform Infrared (FTIR) spectroscopy enable real-time allergen monitoring without altering food integrity, offering potential for inline processing control [6]. Additionally, cloud-based allergen management systems that integrate multiple data streams (ATP monitoring, allergen test results, sanitation records) provide comprehensive risk assessment tools for food manufacturers operating with complex product portfolios [6].
The international shift toward protein-based quantification in programs like VITAL (Voluntary Incidental Trace Allergen Labeling) reflects growing recognition that reporting results in "ppm protein" rather than "ppm matrix" provides more accurate risk assessment for allergic consumers [46]. This transition acknowledges that allergenic reactions are triggered by specific proteins rather than the total mass of allergenic food, emphasizing the importance of method selection in protection of public health [46].
The comparative analysis of protein-based versus DNA-based allergen detection methods reveals a complex landscape where matrix effects significantly influence methodological performance. Protein-based methods, particularly LC-MS/MS, provide direct measurement of immunologically relevant components and demonstrate superior specificity in discriminating between cross-reactive allergens. DNA-based methods offer advantages in analyzing highly processed foods where protein integrity is compromised but cannot directly quantify allergenic risk. The selection of an appropriate detection strategy must consider the specific matrix composition, processing history, and analytical objectives, with emerging technologies promising enhanced capabilities for overcoming matrix interference in complex food systems.
In the comparative analysis of protein-based versus DNA-based detection methods for food allergens, two fundamental specificity issues emerge as critical to understanding performance: antibody cross-reactivity in protein-based immunoassays and false positives in DNA-based polymerase chain reaction (PCR) techniques. Antibody cross-reactivity occurs when antibodies directed against a specific target antigen also bind to structurally similar, but distinct, epitopes on different proteins. This remains a significant challenge for protein-based methods like Enzyme-Linked Immunosorbent Assay (ELISA), which rely on antibody-antigen recognition [51]. Conversely, DNA-based methods like PCR, while offering advantages in stability and specificity for certain applications, face their own false positive challenges primarily from contamination and amplification artifacts [52] [53].
The clinical and diagnostic implications of these specificity issues are substantial. Cross-reactive antibodies can lead to misdiagnosis, unnecessary avoidance of safe foods, and inaccurate epidemiological data. Similarly, false positive PCR results can trigger unnecessary product recalls, economic losses, and erode confidence in food safety testing protocols. This analysis systematically compares these specificity challenges through experimental data, methodological protocols, and mechanistic insights to inform method selection and development for researchers, scientists, and drug development professionals.
Antibody Cross-Reactivity: Mechanisms and Experimental Evidence
Molecular Mechanisms of Antibody Cross-Reactivity
Cross-reactivity stems from the fundamental nature of antibody-antigen interactions. Antibodies recognize specific molecular structures (epitopes) on antigens. When similar epitopes exist on different proteins, a single antibody can bind to multiple targets. This occurs through two primary mechanisms: shared linear epitopes (identical amino acid sequences) and conformational mimicry (structurally similar three-dimensional surfaces) [54]. The degree of cross-reactivity depends on epitope similarity, antibody affinity, and assay conditions.
In food allergy diagnostics, cross-reactivity is particularly problematic within botanical families or between related animal species. For example, antibodies targeting seed storage proteins in peanuts may cross-react with similar proteins in other legumes. Pre-existing immune responses to common viruses or other environmental antigens can also generate cross-reactive antibodies that interfere with specific pathogen diagnostics, as documented between dengue virus and SARS-CoV-2 serological tests [51].
Experimental Evidence and Prevalence Data
A 2022 study systematically evaluated cross-reactivity in anti-SARS-CoV-2 antibody tests using pre-pandemic sera from patients with tropical diseases, providing controlled evidence of this phenomenon. The key findings are summarized in Table 1 below.
Table 1: False Positive Rates in SARS-CoV-2 Serology Across Patient Groups [51]
| Patient Group | Sample Size | Overall False Positive Rate | IgA False Positives | IgM False Positives | IgG False Positives |
|---|---|---|---|---|---|
| All Non-COVID-19 Samples | 170 | 27 (15.9%) | 18 (10.6%) | 9 (5.3%) | 3 (1.8%) |
| Adult Dengue Patients | 80 | Not specified | 11.3% | 5.0% | Not specified |
| Other Tropical Diseases | 30 | Not specified | 16.7% | 13.3% | Not specified |
Notably, the study found that cross-reactive antibodies were most prevalent in the IgA and IgM classes, suggesting these isotypes may have broader binding specificities. The researchers implemented a urea dissociation method to mitigate false positivity, which significantly reduced both false and true positive signals by dissociating low-avidity antibody-antigen complexes [51]. This methodological adjustment demonstrates how understanding cross-reactivity mechanisms can lead to improved assay specificity.
Beyond infectious disease diagnostics, protein-based food allergen detection kits frequently exhibit cross-reactivity with related species. For instance, celery allergen detection may cross-react with other members of the Apiaceae family like carrot and parsley, complicating accurate identification [5]. This highlights a fundamental limitation of protein-based immunoassays when analyzing complex food matrices containing phylogenetically related ingredients.
Despite high theoretical specificity, PCR tests in practice encounter multiple sources of false positives. Contamination represents the most significant challenge, particularly given the exponential amplification nature of PCR that can detect minute quantities of non-target DNA. The primary sources of contamination include:
- Carryover contamination: Amplified PCR products from previous reactions contaminating new samples [52]
- Cross-contamination during sample processing: Improper handling between samples [52]
- Reagent contamination: Manufacturing issues or laboratory handling introducing target sequences [52]
- Cross-reactivity with related genetic sequences: Primers binding to non-target DNA with similar sequences [53]
Additional technical issues contributing to false positives include sample mix-ups, software interpretation errors, and problematic threshold setting for indeterminate results, particularly with low viral loads or DNA concentrations [52].
Quantitative Impact and Predictive Values
The clinical impact of PCR false positives becomes particularly pronounced in low-prevalence settings. As shown in Table 2, even tests with high specificity can produce concerning numbers of false positive results when the target condition is rare in the population being tested.
Table 2: Impact of False Positive Rates on Test Reliability in Low-Prevalence Settings [52]
| Testing Scenario | Prevalence | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| Diagnostic Setting | 10% | 95% | 95% | 84% | Not specified |
| Screening Setting | 1% | 95% | 98% | 32.4% | Not specified |
| Ultra-Low Prevalence | 0.1% | 95% | 98% | 4.5% | Not specified |
External quality assessments of COVID-19 PCR tests revealed false positive rates ranging from <0.4% to 0.7%, with a pooled mean of 0.6% [53]. While these percentages appear small, their impact magnifies substantially in mass screening environments. For example, in a workplace screening 10,000 employees with 1% true infection prevalence, a test with 95% sensitivity and 98% specificity would identify 95 true positives but also 198 false positives, meaning nearly two-thirds of all positive results would be incorrect [52].
Real-world data from The Walt Disney Company's testing program during COVID-19 documented that 22.6% of positive tests in their screening program were ultimately determined to be false positives after retesting, highlighting the practical significance of this issue [52].
Comparative Methodologies: Experimental Protocols
Urea Dissociation Protocol for Reducing Antibody Cross-Reactivity
The urea dissociation method provides a biochemical approach to distinguish high-affinity specific antibodies from low-affinity cross-reactive binders. The detailed protocol, as applied to SARS-CoV-2 ELISA, involves these critical steps [51]:
- Coating: Immobilize antigens (spike S1 or nucleocapsid proteins) onto microplates.
- Sample Incubation: Add diluted patient serum (1:100 in sample buffer) and incubate to allow antibody-antigen binding.
- Urea Treatment: Add 100 µL of PBS solution containing 4 mol/L urea and incubate at 37°C for 10 minutes. This concentration disrupts hydrogen bonds, dissociating low-avidity interactions.
- Washing: Remove urea and dissociated antibodies through standard washing procedures.
- Detection: Proceed with enzyme-conjugated secondary antibodies and substrate addition per standard ELISA protocol.
- Measurement: Read optical density at 450 nm and calculate ratios against calibrators.
This method capitalizes on the principle that cross-reactive antibodies typically form lower avidity interactions that are more susceptible to disruption by chaotropic agents like urea. Implementation in the tropical disease study significantly reduced false positive signals, demonstrating its utility for improving serological assay specificity [51].
DNA Extraction and qPCR Protocol for Allergen Detection
For DNA-based allergen detection, the following protocol adapted from celery detection studies provides a framework for minimizing false positives [5]:
DNA Extraction:
- Weigh ~30 mg of sample material into a 2 mL tube.
- Add 1 mL of CTAB buffer, 40 μL of proteinase K (20 mg/mL), and 5 μL of RNase (100 mg/mL).
- Incubate for 90 minutes at 65°C.
- Centrifuge for 10 minutes at 16,000×g.
- Transfer 300 μL of supernatant to a Maxwell cartridge.
- Purify using Maxwell RSC 48 instrument with PureFood GMO and authentication method.
- Elute DNA in 100 μL elution buffer.
qPCR Setup:
- Dilute DNA to 10 ng/μL concentration.
- Combine 5 μL DNA with 20 μL reaction mix containing:
- TaqMan Universal Master Mix
- Target-specific primers (300 nmol/L final concentration)
- Probe (200 nmol/L final concentration)
- For celery detection, target the mannitol dehydrogenase gene with:
- Forward primer: CGATGAGCGTGTACTGAGTC
- Reverse primer: AATAGGAACTAACATTAATCATACCAAAC
- Probe: FAM-AACAGATAACGCTGACTCATCACACCG-BBQ
Amplification:
- Run on CFX-96 instrument with protocol:
- 2 minutes at 50°C
- 10 minutes at 95°C
- 45 cycles of: 15 seconds at 95°C, 1 minute at 60°C
- Run on CFX-96 instrument with protocol:
Contamination Prevention Measures:
- Physical separation of pre- and post-amplification areas
- Use of dedicated equipment and consumables
- Implementation of negative controls in each run
- Careful threshold setting to distinguish true positives from background
This protocol highlights the importance of both biochemical optimization and procedural controls to minimize false positives in DNA-based detection [5].
Visualization of Key Concepts and Workflows
Antibody Cross-Reactivity Mechanisms
Figure 1: Antibody Cross-Reactivity Mechanisms
PCR Contamination Pathways
Figure 2: PCR False Positive Pathways
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Specificity Optimization
| Reagent / Material | Application | Function in Specificity Optimization |
|---|---|---|
| Urea (4 mol/L solution) | Protein-based assays | Chaotropic agent that dissociates low-avidity antibody-antigen complexes to reduce cross-reactivity [51] |
| CTAB Buffer | DNA extraction | Surfactant that facilitates cell lysis and stabilizes nucleic acids during extraction from complex matrices [5] |
| Proteinase K | DNA extraction | Proteolytic enzyme that degrades nucleases and proteins, improving DNA yield and purity [5] |
| TaqMan Probes | qPCR assays | Fluorogenic probes that increase specificity through dual recognition (primers + probe) requiring three specific binding events for signal generation [5] |
| Rheumatoid Factor Absorbent | Serological assays | Removes rheumatoid factor and other interfering antibodies that cause false positives in immunoassays [55] |
| Maxwell RSC System | Automated nucleic acid purification | Standardized extraction platform that reduces cross-contamination between samples compared to manual methods [5] |
The comparative analysis of specificity challenges in protein-based versus DNA-based detection methods reveals distinctive advantage and limitation profiles. Protein-based immunoassays face fundamental challenges with antibody cross-reactivity, particularly when analyzing samples containing phylogenetically related proteins or from individuals with pre-existing immunological conditions. The urea dissociation method and other avidity-based techniques offer promising approaches to mitigate these limitations.
DNA-based PCR methods, while theoretically highly specific, confront practical false positive challenges primarily from contamination and amplification artifacts. Rigorous laboratory practices, spatial separation of pre- and post-amplification areas, and careful threshold setting are essential for maintaining specificity.
Method selection should be guided by the specific application context: protein-based methods may be preferred when assessing functional allergenicity or protein presence, while DNA-based approaches offer advantages for species identification in processed foods where DNA stability exceeds protein stability. Future directions should focus on orthogonal testing approaches that combine both methodologies, advanced bioinformatic tools for epitope and primer prediction to minimize cross-reactivity, and continued refinement of extraction and amplification protocols to reduce contamination vulnerabilities.
For researchers and drug development professionals, this analysis underscores that both methodological approaches require sophisticated implementation and validation strategies to overcome their characteristic specificity challenges. Understanding these fundamental mechanisms enables more informed method selection, protocol optimization, and interpretation of diagnostic results across food safety, clinical diagnostics, and biomedical research applications.
Optimization Strategies for Sample Extraction, Purification, and Result Standardization
Food allergy is a significant global public health issue, for which strict avoidance of allergenic foods remains the most effective preventive measure. Accurate food allergen detection is therefore critical for consumer protection. The two predominant analytical approaches are protein-based methods (e.g., ELISA, Mass Spectrometry) and DNA-based methods (e.g., PCR, isothermal amplification). The performance of these methods is profoundly influenced by the initial steps of sample preparation, including extraction and purification. The efficacy of allergen detection in food products is often limited by complex matrix effects, the presence of interfering compounds, and the impact of food processing on analyte integrity. This guide provides a comparative analysis of optimization strategies for sample preparation and standardization of results, contextualized within the broader comparison of protein-based and DNA-based allergen detection methodologies [1] [56] [57].
Comparative Analysis of Allergen Detection Methods
Fundamental Principles and Targets
- Protein-Based Methods: These methods directly detect the allergenic proteins themselves. Immunoassays like ELISA use antibodies to recognize and bind specific protein epitopes, while Mass Spectrometry (MS) identifies unique peptide sequences derived from allergenic proteins. Their performance is directly affected by protein integrity, which can be compromised by food processing that denatures proteins and alters antibody-binding sites [1] [56].
- DNA-Based Methods: These are indirect methods that detect DNA sequences (genes) encoding the allergenic proteins or other specific genomic markers. Techniques like PCR amplify specific DNA fragments for detection. DNA is generally more stable than proteins during thermal processing, making these methods particularly suitable for detecting allergens in highly processed foods where proteins may have become denatured [1] [56] [5].
Performance Comparison Table
The following table summarizes the key characteristics of the two detection approaches, highlighting how their differing targets and principles lead to distinct advantages and limitations [1] [56] [57].
Table 1: Comparative Analysis of Protein-based and DNA-based Allergen Detection Methods
| Characteristic | Protein-Based Methods | DNA-Based Methods |
|---|---|---|
| Analytical Target | Allergenic proteins or peptides | DNA encoding allergenic proteins or specific genomic markers |
| Primary Techniques | ELISA, Lateral Flow Immunoassay (LFI), Mass Spectrometry | PCR (qPCR, digital PCR), Isothermal Amplification, Biosensors |
| Impact of Food Processing | High susceptibility; protein denaturation can lead to false negatives | Lower susceptibility; DNA is more thermally stable, but fragmentation can occur |
| Specificity | High, but potential for cross-reactivity with related proteins | High, with ability to discriminate between closely related species (e.g., celery vs. carrot) |
| Sensitivity | High (e.g., ELISA can detect 1-5 ppm of peanut protein) | High (e.g., can detect down to 1 ppm celery DNA in spiked foods) |
| Quantification | Direct quantification of the allergenic component | Indirect quantification; correlation between DNA amount and protein content can be variable |
| Matrix Interference | Susceptible to interference from polyphenols (e.g., in chocolate), fats, and other proteins | Susceptible to interference from polysaccharides, polyphenols, and other PCR inhibitors |
| Key Challenge | Developing antibodies that are resilient to processing-induced protein changes | Selecting appropriate DNA targets and converting DNA results to accurate protein estimates |
Optimization of Sample Preparation
The accuracy of any allergen detection method is contingent on the efficient and reproducible recovery of the target analyte from the food matrix. Sample preparation is therefore a critical step.
Key Considerations for Extraction
- Matrix Composition: The food matrix itself is a major source of interference. Ingredients like polyphenols (common in chocolate and spices), fats, and salts can bind to or co-extract with the target, inhibiting subsequent antibody binding in immunoassays or enzymatic reactions in PCR [57] [5].
- Food Processing Effects: Thermal processing (e.g., baking, frying) can denature proteins, making them unrecognizable to antibodies. While more stable, DNA can also be fragmented by high temperatures, requiring the design of PCR assays that amplify short DNA fragments (typically 200-300 bp) for reliable detection in processed foods [57] [7].
- Allergen Source Material: Different parts of a plant (e.g., celery root vs. celery stem) can have varying protein and DNA content, extractability, and stability, which must be considered during method development and validation [5].
Optimized Extraction Buffers and Additives
Research has identified that the composition of the extraction buffer is paramount for maximizing analyte recovery. A systematic study comparing 10 different buffers for extracting 14 different allergens from complex matrices like chocolate dessert and baked biscuits identified two optimized buffer formulations [57]:
- Buffer D: 50 mM carbonate bicarbonate with 10% fish gelatine, pH 9.6
- Buffer J: PBS with 2% Tween-20, 1 M NaCl, 10% fish gelatine, and 1% Polyvinylpyrrolidone (PVP), pH 7.4
These buffers, incorporating specific additives, were found to provide recoveries of 50-150% for most allergens in the tested matrices, though challenging matrices like chocolate and thermally processed foods still showed lower recoveries [57].
Table 2: Key Buffer Additives and Their Functions in Allergen Extraction
| Additive | Function | Applicability |
|---|---|---|
| Fish Gelatine (10%) | Protein-blocking agent; reduces non-specific binding and protein adsorption to surfaces. | Protein-based & DNA-based methods |
| PVP (1%) | Binds and removes polyphenols, preventing them from inhibiting assays. | Protein-based & DNA-based methods (especially for polyphenol-rich matrices) |
| High Salt (1 M NaCl) | Increases ionic strength to disrupt protein-matrix and DNA-matrix interactions. | Protein-based & DNA-based methods |
| Detergent (2% Tween-20) | Solubilizes proteins and lipids, facilitating the release of targets from the matrix. | Protein-based & DNA-based methods |
| SDS (1%) | Strong ionic detergent that denatures proteins and aids in solubilization. | Protein-based methods (can interfere with some immunoassays) |
| Sodium Sulphite | A reducing agent that can help break disulfide bonds in proteins, aiding extraction. | Protein-based methods |
Experimental Protocol: Optimized Allergen Extraction
This protocol is adapted from studies that successfully extracted multiple allergens from complex, incurred food matrices for immunoassay analysis [57].
Methodology:
- Homogenization: Weigh 1 g of the representative food sample. For solid foods, homogenize using a mortar and pestle or a mechanical grinder until a fine, consistent powder is achieved.
- Extraction: Add the sample to 10 mL of a pre-warmed optimized extraction buffer (e.g., Buffer D or J from Table 2) at a 1:10 (w/v) ratio.
- Incubation: Vortex mix for 30 seconds to ensure thorough suspension. Incubate the mixture for 15 minutes in an orbital shaker incubator set to 60°C and 175 rpm. The elevated temperature and agitation aid in the efficient release of the target analytes.
- Clarification: Centrifuge the extract at 1250-1600 x g for 20 minutes at 4°C to pellet insoluble debris.
- Collection: Carefully collect the clarified supernatant from the middle of the tube, avoiding the surface lipid layer and the bottom pellet, for subsequent analysis.
Standardization and Data Interpretation
Quantification and Reference Materials
A significant challenge in allergen detection is the accurate quantification and interpretation of results.
- Protein-Based Methods: ELISA kits are typically calibrated using crude extracts of the allergenic source (e.g., peanut flour), leading to potential ambiguity in the exact protein being quantified and a lack of harmonization between different kits [57]. A move towards allergen-specific immunoassays, which use purified, clinically relevant allergen proteins for calibration, offers improved standardization and reporting clarity [57].
- DNA-Based Methods: Quantification is inherently indirect. While qPCR can precisely measure the amount of a specific DNA sequence, converting this DNA concentration into the amount of allergenic protein present is challenging. Factors such as gene copy number variation, differences between plant tissues, and the effects of processing can lead to an overestimation or underestimation of the actual allergen protein content [5]. The use of an internal amplification control (IAC) is crucial in qPCR to identify the presence of PCR inhibitors in the sample, which could otherwise cause false negatives [56].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Allergen Detection Development
| Reagent / Material | Function | Application Context |
|---|---|---|
| Fish Gelatine | Non-specific blocking agent in extraction buffers. | Reduces surface adsorption of proteins, improving recovery in immunoassays and DNA extraction [57]. |
| Polyvinylpyrrolidone (PVP) | Binds and precipitates polyphenols. | Critical for extracting allergens from chocolate, spices, and other polyphenol-rich matrices [57]. |
| CTAB Buffer | Cetyltrimethyl ammonium bromide buffer for DNA extraction. | Effectively co-precipitates polysaccharides; ideal for DNA isolation from complex plant-based matrices [7]. |
| Proteinase K | Broad-spectrum serine protease. | Digests contaminating proteins and nucleases during DNA extraction, protecting the target DNA [5]. |
| TaqMan Probes | Hydrolysis probes for qPCR. | Provide sequence-specific detection, enabling highly specific and quantitative DNA analysis [5]. |
| Allergen-Specific Antibodies | Monoclonal or polyclonal antibodies. | Key reagents for specific immunoassays (ELISA, LFD) targeting individual allergenic proteins [57]. |
| Molecularly Imprinted Polymers (MIPs) | Synthetic polymer receptors with tailor-made recognition sites. | Used in biosensors as stable, cost-effective alternatives to antibodies for detecting small molecule allergen tracers [45]. |
Visualizing Allergen Detection Workflows
The following diagrams illustrate the core workflows for protein-based and DNA-based allergen detection, highlighting the critical steps of sample preparation.
Figure 1: Protein-Based Allergen Detection Workflow. This process involves extracting proteins, often with specialized buffers, before immunoassay or mass spectrometric analysis.
Figure 2: DNA-Based Allergen Detection Workflow. This process focuses on isolating and purifying DNA, followed by amplification of specific marker genes.
A Rigorous Comparative Framework: Sensitivity, Specificity, and Applicability
The accurate detection of food allergens is a critical component of public health strategies, necessitating analytical methods that are both highly sensitive and reliable. The detection landscape is predominantly divided into two methodological approaches: those that target allergenic proteins directly and those that target the DNA of the allergenic source. Protein-based methods, such as Enzyme-Linked Immunosorbent Assay (ELISA) and mass spectrometry, detect the very molecules responsible for eliciting an immune response. In contrast, DNA-based methods, primarily polymerase chain reaction (PCR) and its variants, detect genetic markers as a surrogate for the presence of the allergenic ingredient [58] [59].
This guide provides a direct performance comparison of these two foundational approaches, focusing on the core analytical parameters of Limits of Detection (LOD), Quantification, and Dynamic Range. For researchers and scientists in drug development and food safety, selecting the appropriate method is paramount. The choice hinges on a clear understanding of these performance characteristics within the context of complex food matrices and varied processing conditions. This analysis synthesizes experimental data from recent, rigorous studies to offer an objective comparison and support informed methodological selection.
Performance Metrics Comparison
The following tables consolidate quantitative performance data for various protein- and DNA-based allergen detection methods as reported in recent scientific literature.
Table 1: Performance Comparison of DNA-Based Allergen Detection Methods
| Allergen | Technology | Target Gene | LOD / LOQ | Dynamic Range | Sample Matrix | Citation |
|---|---|---|---|---|---|---|
| Sesame | Nanoplate Digital PCR (ndPCR) | ITS | 0.1 mg/kg | Not Specified | Dough, Biscuits | [60] |
| Sesame | Nanoplate Digital PCR (ndPCR) | CO6b-1 | 5 mg/kg | Not Specified | Dough, Biscuits | [60] |
| Soy | LAMP-LFD | Lectin | 0.005% (50 mg/kg) | Not Specified | Various Food Samples | [61] [59] |
| Soy | LAMP-LFD | ORF160b | 10 mg/kg | Not Specified | Boiled Sausage, Chocolate, Soup | [59] |
| Sesame | Real-time PCR | 2S Albumin | 0.005% (50 mg/kg) | Not Specified | Various Food Samples | [59] |
| Pistachio | Real-time PCR | 2S Albumin | 0.004% (40 mg/kg) | Not Specified | Various Food Samples | [59] |
| Macadamia | Real-time PCR | Vicilin Precursor | 0.006% (60 mg/kg) | Not Specified | Various Food Samples | [59] |
| Peanut | Real-time PCR | Chloroplast matK | 10 mg/kg | Adequate Efficiency & Linearity | Processed Samples | [59] |
| Gluten, Sesame, Soy, Hazelnut | Microfluidic qPCR | Species-Specific | Meets 20 ppm Gluten Threshold | Not Specified | Flour, Spread, Canned Food | [58] |
Table 2: Performance and Characteristics of Protein-Based Allergen Detection Methods
| Allergen | Technology | Detected Molecule | LOD / LOQ | Key Characteristics & Challenges | Citation |
|---|---|---|---|---|---|
| Gluten | ELISA (R5 Antibody) | Gliadins/Gluetnins | 2 ppm (Sensitivity) | "Gold standard" for gluten; Dynamic range of 10-30 ppm demonstrated in microfluidic immunoassay. | [58] [59] |
| Pistachio | Phage-Display ELISA | Pis v 2 (11S Globulin) | ~4000 mg/kg | Specific to pistachio; no cross-reactivity with cashew. | [59] |
| Seafood | LC-MS/MS (Targeted) | Peptide Biomarkers | Analysis in <2 hours | High specificity; overcomes antibody limitations and epitope masking. | [62] [59] |
| Various | Nanozyme Biosensors | Allergenic Proteins | High Sensitivity (Theoretical) | Exceptional storage stability and reusability; emerging technology. | [63] |
Experimental Protocols for Key Studies
Digital PCR for Sesame Detection
This protocol is adapted from the study that established the first nanoplate digital PCR (ndPCR) method for tracing allergenic foods, demonstrating superior sensitivity for sesame [60].
- 1. Sample Preparation: Food samples (e.g., dough, biscuits) were homogenized. A representative subsample was used for DNA extraction.
- 2. DNA Extraction: Genomic DNA was purified from 100 mg of the homogenized sample using a CTAB-based method. The method involved incubation with CTAB buffer and proteinase K at 65°C, RNase A treatment, chloroform extractions, and DNA precipitation with isopropanol [7]. DNA concentration and purity were assessed using UV-Vis spectrophotometry.
- 3. Primer Design: Two independent ndPCR assays were designed targeting:
- The CO6b-1 region (single-copy gene).
- The Internal Transcribed Spacer (ITS) region (multi-copy gene).
- 4. Nanoplate Digital PCR Setup: The ndPCR reaction mix was prepared using a mastermix suitable for digital PCR. The mixture was loaded into a nanoplate, which partitions the sample into thousands of individual nanoreactions.
- 5. Amplification: The nanoplate was cycled in a thermal cycler with optimized annealing temperatures for each primer set.
- 6. Data Analysis: After amplification, each nanoreaction was analyzed for fluorescence. Partitions containing the target sequence were scored as positive. The absolute quantity of target DNA in the original sample was calculated using Poisson statistics.
LAMP-LFD for Soy Detection
This protocol details the Loop-Mediated Isothermal Amplification combined with a Lateral Flow Device (LAMP-LFD) method, which offers a rapid and low-technology alternative for soybean detection [61] [59].
- 1. DNA Extraction: Genomic DNA from food matrices (e.g., boiled sausage, chocolate, instant tomato soup) was obtained using a commercial plant DNA extraction kit, following the manufacturer's instructions.
- 2. LAMP Primer Design: LAMP primers (inner and outer primers) were designed to target a multicopy gene (ORF160b) for high sensitivity.
- 3. LAMP Reaction: The reaction was carried out in an isothermal mastermix at a constant temperature (60–65 °C) for 30-60 minutes. The forward inner primer (FIP) was labeled with biotin.
- 4. Lateral Flow Detection: After amplification, the LAMP product was applied to the LFD. Gold nanoparticles conjugated with an antibody (e.g., anti-FITC) bind to the labeled amplicon. This complex migrates and is trapped at a test line, producing a visible band.
- 5. Integrated Quality Control: A second LAMP assay targeting a housekeeping gene (e.g., Cytochrome Oxidase, Cox) was implemented. Its FIP was labeled with a different hapten (e.g., digoxin), allowing a separate control line on the LFD to verify successful DNA extraction and amplification, thus mitigating false negatives.
- 6. Result Interpretation: Results were interpreted visually or using a digital cube reader for data traceability and sharing.
Integrated Microfluidic qPCR for Multiplex Allergen Detection
This protocol describes a fully automated "sample-to-result" platform for the simultaneous detection of four allergens [58].
- 1. Sample Loading: Several grams of a complex food matrix (flour, spread, seed mix) were loaded into a dedicated microfluidic cartridge.
- 2. Automated Sample Preparation (On-Cartridge):
- Extraction & Concentration: Integrated pumps, valves, and a filter membrane autonomously performed DNA extraction, concentration, and purification from a large sample volume to ensure representativity.
- Purification: The process removed PCR inhibitors commonly found in complex matrices (e.g., fats, polyphenols).
- 3. Multiplex qPCR (On-Cartridge): The purified DNA was automatically distributed into multiple reaction chambers pre-loaded with primers and probes for:
- Target Allergens: Gluten, sesame, soy, and hazelnut.
- Controls: A negative control chamber and an internal positive control in each chamber to validate results.
- 4. Instrument Operation & Data Analysis: The in-house instrument provided pneumatic control for fluidics, thermal control for qPCR, and optical detection for multiplex fluorescence. Data analysis software provided quantification cycle (Cq) values, ensuring results were protected from false positives/negatives.
Visualized Workflows and Signaling Pathways
DNA-Based Allergen Detection Workflow
The following diagram illustrates the generalized workflow for DNA-based allergen detection methods, from sample preparation to final result interpretation.
Protein-Based Allergen Detection Workflow
The following diagram illustrates the core principles of protein-based detection, highlighting both immunoassay and mass spectrometry pathways.
IgE-Mediated Allergic Reaction Pathway
Understanding the mechanism of IgE-mediated allergies, which protein-based methods directly assess, is crucial for contextualizing their detection goals. The following diagram outlines this immune signaling pathway.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Food Allergen Detection Research
| Item | Function/Application | Specific Examples / Notes | |
|---|---|---|---|
| DNA Extraction Kits | Purification of genomic DNA from complex food matrices. | CTAB-based methods; Commercial kits (e.g., Qiagen DNeasy Plant Mini Kit). | [61] [7] |
| Isothermal Mastermix | Enzymatic mix for LAMP amplification at constant temperature. | Optigene Isothermal Mastermix; Contains strand-displacing DNA polymerase. | [61] |
| qPCR Mastermix | Enzymatic mix for real-time PCR, containing polymerase, dNTPs, and buffer. | Requires thermostable polymerase (e.g., Taq); May include UNG to prevent carryover contamination. | [58] |
| Lateral Flow Devices (LFD) | Rapid, visual detection of labeled amplicons or proteins. | Milenia HybriDetect 2T; Contains test and control lines with immobilized antibodies. | [61] |
| Microfluidic Cartridges | Integrated platforms for automated sample preparation and analysis. | Custom-designed cartridges with pumps, valves, and reaction chambers. | [58] |
| Specific Antibodies | Core recognition element in immunoassays for allergenic proteins. | R5 monoclonal antibody for gluten; PVF4 phage-dAb for pistachio Pis v 2. | [59] |
| Mass Spectrometry Standards | Synthetic isotope-labeled peptides for absolute quantification (LC-MS/MS). | AQUA peptides used in selected/multiple reaction monitoring (SRM/MRM). | [62] [59] |
| Nanozymes | Engineered nanomaterial catalysts mimicking enzyme activity in biosensors. | Potential alternative to natural enzymes with superior stability; used in electrochemical/optical sensors. | [63] |
Analysis of Cost, Throughput, and Ease of Use for Laboratory vs. On-Site Testing
Within food safety and public health, the accurate detection of food allergens is paramount. As the incidence of food allergies continues to rise globally, the demand for reliable, efficient, and accessible testing methods has intensified [3]. This comparison guide objectively analyzes two fundamental testing paradigms: centralized laboratory testing and on-site (point-of-care) testing. The analysis is specifically framed within ongoing research comparing protein-based and DNA-based food allergen detection methods. The choice between laboratory and on-site testing influences cost, throughput, and ease of use, thereby impacting the efficiency of research and the development of new diagnostic tools and food safety protocols. This guide provides a structured comparison to assist researchers, scientists, and drug development professionals in selecting the optimal testing approach for their specific applications, particularly when evaluating the efficacy of protein versus DNA methodologies.
Comparative Analysis of Testing Modalities
The decision between laboratory and on-site testing involves a trade-off between multiple performance factors. The following sections provide a detailed comparison of these two modalities.
Cost Analysis
Cost structures differ significantly between laboratory and on-site testing. Centralized laboratories benefit from economies of scale, leading to a lower per-test cost for the actual analysis [64]. However, this does not always translate to lower costs for the end user. A 2024 study comparing physician-ordered hospital laboratory tests with Direct-to-Consumer (DTC) tests, which often utilize major laboratory networks, found that DTC prices were generally lower than the mean insurance-negotiated hospital-based prices [65]. For uninsured patients, the savings were even more substantial.
Table 1: Cost Comparison for Common Laboratory Tests (Data from [65])
| Laboratory Test | Hospital Outpatient Charge (Mean $) | Hospital Insurance-Negotiated Price (Mean $) | Direct-to-Consumer / On-Site Cost (Mean $) |
|---|---|---|---|
| Complete Blood Count | 401.00 | 88.00 | 32.00 |
| Hemoglobin A1c | 250.00 | 81.00 | 42.00 |
| Vitamin D | 173.00 | 72.00 | 87.00 |
| Metabolic Profile | 957.00 | 218.00 | 52.00 |
| Lipid Profile | 558.00 | 110.00 | 62.00 |
| Prostate-Specific Antigen | 276.00 | 83.00 | 72.00 |
Furthermore, a report from the Health Care Cost Institute (HCCI) highlighted that employer-sponsored insurance pays three to six times more for the same lab test when performed by a hospital outpatient lab compared to a physician's office or an independent lab [66]. For example, a basic metabolic panel cost $7.75 in an office/independent lab setting versus $38.44 in a hospital outpatient setting. These cost disparities are significant for research budgeting and development of cost-effective testing solutions.
For on-site testing, the cost structure includes upfront capital investment in portable instruments and ongoing costs for cartridges or test strips [67]. While the per-test consumable cost may be higher, on-site testing eliminates sample transport logistics and can reduce overall project costs by enabling immediate decision-making [68].
Throughput and Turnaround Time
Throughput and turnaround time are critical factors where the two modalities exhibit clear, opposing strengths.
Table 2: Throughput and Turnaround Time Comparison
| Feature | Laboratory Testing | On-Site Testing |
|---|---|---|
| Testing Speed | Time-intensive due to transport, setup, and queuing [69] | Immediate results; tests can be run within minutes or hours [68] |
| Result Delivery | Delays possible due to transport and processing time [69] | Rapid delivery of results to practitioners on-site [67] |
| Batch Processing | High; optimized for processing a large volume of samples efficiently [70] | Low to moderate; typically limited by the number of cartridge ports on the device [67] |
| Impact on Workflow | Delays in result reporting can slow research and patient management [64] | Faster diagnosis and treatment; enables quick adjustments in experimental protocols [68] |
Central laboratories are designed for high-throughput analysis, processing large batches of samples with sophisticated, automated instruments [70]. This comes at the expense of time, as transport to the facility adds significantly to the total turnaround time [64]. In contrast, on-site testing provides rapid turnaround, offering results within the same clinical or experimental session. This speed is its primary advantage, minimizing the risk of losing patients to follow-up in clinical settings and allowing for rapid iteration in research [64] [67].
Ease of Use and Control
Ease of use encompasses the complexity of operation, the level of training required, and the degree of control over the testing environment and process.
Table 3: Ease of Use and Operational Control Comparison
| Feature | Laboratory Testing | On-Site Testing |
|---|---|---|
| Operator Skill | Requires specialized staff and training [64] | Designed for ease of use, often by non-specialist staff [67] |
| Environmental Control | Highly controlled environment minimizes interference [69] | Real-world conditions with variables like temperature and humidity [71] |
| Process Control | Standardized protocols and rigorous quality control [70] | Greater direct control over the testing process and investigation of discrepancies [68] |
| Communication | Potential for communication delays between sites [70] | Improved communication between doctors and technicians on-site [68] |
| Test Menu & Flexibility | Access to a wide range of specialized and complex tests [70] | Limited to available test cartridges; ideal for specific, routine tests [68] |
Laboratory testing occurs in a controlled environment, which minimizes external variables and ensures the consistency and reproducibility of results [69]. The procedures are highly standardized, and these facilities undergo regular external accreditation, ensuring adherence to strict quality standards like Good Laboratory Practice (GLP) [70]. However, this requires highly trained personnel.
On-site testing prioritizes accessibility and convenience. The instruments are often designed for simplicity, potentially enabling task-shifting to nurses or other non-laboratory staff in a clinical trial context [67]. This provides greater immediate control over the testing process and allows for direct investigation of any anomalies [68]. The trade-off is that testing is performed in a variable environment, which can influence accuracy, and the range of available tests is typically limited to the device's predefined cartridges [69] [68].
Decision Framework for Testing Selection
The choice between laboratory and on-site testing is not universal but should be based on the specific requirements of the project, the characteristics of the disease or analyte, and the available resources. A generalized decision-making framework can be visualized as follows, highlighting the key trade-offs.
The most critical determinants in this framework are the clinical or experimental utility lost due to send-out delays and the relative accuracy of onsite versus laboratory testing [64]. Modeling suggests that when the sensitivity of on-site testing is high, it is preferred when delays in laboratory testing would reduce clinical utility by more than 20%. However, if on-site testing incurs a large reduction in sensitivity, it may only be preferable when the utility lost due to delays exceeds 50% [64]. The relative cost of testing also impacts these thresholds, particularly when the test cost is significant relative to the cost of treatment or the overall project budget.
Application in Food Allergen Detection Research
The comparison between laboratory and on-site testing is highly relevant in the context of research comparing protein-based and DNA-based food allergen detection methods. Each detection method has inherent characteristics that may align better with one testing modality.
Protein-Based Detection Methods
Experimental Protocols: Protein-based methods, such as Enzyme-Linked Immunosorbent Assay (ELISA) and Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS), directly detect allergenic proteins [1] [3]. ELISA works by immobilizing food samples on a plate, then using antibodies specific to the allergen protein that are linked to an enzyme. A substrate is added, producing a colorimetric signal measured spectrophotometrically [1]. LC-MS/MS involves digesting proteins into peptides, separating them by liquid chromatography, and identifying them based on their mass-to-charge ratio [3].
- Alignment with Testing Modality: These methods, especially LC-MS/MS, typically require sophisticated, non-portable instrumentation and a controlled environment for optimal performance [1]. They are, therefore, predominantly performed in central laboratories. The CAC has adopted ELISA as an official test for gluten allergens, underscoring its role in standardized, lab-based settings [1].
DNA-Based Detection Methods
Experimental Protocols: DNA-based methods, primarily the Polymerase Chain Reaction (PCR) and its variants (e.g., real-time PCR), indirectly detect allergens by amplifying unique DNA sequences of the allergenic source [1]. The process involves extracting DNA from a food sample, designing primers specific to the target allergen (e.g., peanut, hazelnut), and amplifying the target sequence through thermal cycling. The amplified product is then detected, often in real-time via fluorescent probes [1].
- Alignment with Testing Modality: DNA is more stable than proteins through various food processing stages, making PCR a robust alternative for processed foods [1]. While PCR is a core technology in central labs, the principles of nucleic acid amplification are the foundation for many rapid, on-site biosensors. Emerging biosensors using microfluidics and isothermal amplification (e.g., loop-mediated isothermal amplification, LAMP) are being developed to integrate DNA extraction, amplification, and detection into portable devices for on-site use [1]. Germany and Japan have established PCR as an official analytical tool for food allergen detection, demonstrating its validity in both lab and potential field applications [1].
The Scientist's Toolkit: Key Research Reagents and Materials
The execution of both protein and DNA-based allergen detection methods relies on a suite of specialized reagents and materials. The following table details key components essential for researchers in this field.
Table 4: Essential Research Reagent Solutions for Allergen Detection
| Reagent / Material | Function in Protein-Based Methods | Function in DNA-Based Methods |
|---|---|---|
| Specific Antibodies | The core reagent in immunoassays (e.g., ELISA); binds specifically to the target allergenic protein epitope to enable detection [1]. | Not typically used. |
| Trypsin/Lys-C Enzymes | Used in LC-MS/MS sample preparation to enzymatically digest proteins into peptides for mass spectrometric analysis [3]. | Not used. |
| DNA Polymerase | Not used. | The key enzyme that synthesizes new DNA strands during PCR amplification; thermostable versions (e.g., Taq polymerase) are essential [1]. |
| Sequence-Specific Primers & Probes | Not used. | Short, single-stranded DNA molecules that are designed to be complementary to and bracket the target allergen gene sequence, enabling specific amplification and detection [1]. |
| Magnetic Beads (functionalized) | Can be coated with antibodies for immunomagnetic separation of allergens from complex food matrices [1]. | Often coated with silica for binding and purifying DNA from complex food samples [1]. |
| Fluorescent Dyes/Reporters | Conjugated to detection antibodies in ELISA to generate a measurable signal [1]. | Incorporated into PCR products (e.g., SYBR Green) or released from hydrolysis probes (e.g., TaqMan) to quantify DNA amplification in real-time [1]. |
| Aptamers | Synthetic single-stranded DNA or RNA oligonucleotides that can bind to specific protein targets; used as antibody alternatives in biosensors [1]. | Can be used in biosensors for detection, but their function is based on protein-binding, not DNA amplification. |
The analysis of cost, throughput, and ease of use reveals that there is no single superior option between laboratory and on-site testing; rather, the optimal choice is context-dependent. Centralized laboratory testing remains the gold standard for high-throughput, standardized, and highly accurate analyses, making it indispensable for specialized assays, regulatory submissions, and large-scale studies. In contrast, on-site testing offers unparalleled advantages in speed, convenience, and the ability to inform immediate decisions, which is critical for rapid diagnostics and iterative research. In the specific field of food allergen detection research, protein-based methods like ELISA and LC-MS/MS are firmly rooted in the laboratory environment due to their instrumentation needs. In comparison, DNA-based methods like PCR, while also a lab staple, provide a pathway toward the development of portable, sensitive, and specific on-site biosensors. Researchers must weigh the trade-offs outlined in this guide—balancing the imperative for speed against the needs for accuracy, cost-control, and standardization—to select the most effective testing modality for their scientific and operational objectives.
Food allergy is a significant global public health concern, affecting nearly 6-8% of children and 3-5% of adults, with its incidence steadily increasing [72]. For sensitized individuals, avoiding allergenic foods remains the primary prevention strategy, as no definitive cure currently exists [1] [72]. This reality places critical importance on accurate allergen detection in food products to protect consumer health and comply with regulatory requirements. The detection of allergens relies primarily on two analytical approaches: protein-based methods that directly detect allergenic proteins, and DNA-based methods that target specific DNA sequences from allergenic sources [1] [56].
Each category of methods possesses distinct advantages and limitations that affect their suitability for different testing scenarios. Protein-based techniques, including immunoassays and mass spectrometry, directly measure the causative agents of allergic reactions. In contrast, DNA-based methods, particularly polymerase chain reaction (PCR), offer an indirect approach that leverages the greater stability of DNA through food processing steps [56]. This comprehensive analysis examines the applicability of these methodological approaches based on food matrix characteristics and testing objectives, providing researchers and food safety professionals with evidence-based guidance for method selection.
Comparative Performance of Allergen Detection Methods
Fundamental Principles and Technical Characteristics
Protein-Based Methods encompass techniques that directly detect allergenic proteins or peptides. Enzyme-Linked Immunosorbent Assay (ELISA) operates on antibody-antigen interactions, providing sensitive and specific quantification of target proteins [1]. Mass Spectrometry (MS), particularly liquid chromatography-tandem mass spectrometry (LC-MS/MS), identifies and quantifies specific allergenic peptides based on their mass-to-charge ratio, offering high specificity and multiplexing capability [50]. Multiplex immunoassays, such as the xMAP Food Allergen Detection Assay, simultaneously detect multiple allergens using antibody-conjugated, color-coded magnetic beads [73].
DNA-Based Methods primarily utilize PCR and its variants to amplify and detect species-specific DNA sequences. Real-time quantitative PCR (qPCR) monitors amplification in real-time, enabling quantification of target DNA [5]. Isothermal amplification techniques, such as loop-mediated isothermal amplification (LAMP), provide rapid detection without thermal cycling [56]. DNA biosensors employ immobilized probes to hybridize with target DNA sequences, often coupled with electrochemical or optical transduction systems [1].
Performance Comparison Across Food Matrices
Table 1: Comparative Analysis of Allergen Detection Methods
| Method | Detection Principle | Sensitivity | Food Matrix Considerations | Advantages | Limitations |
|---|---|---|---|---|---|
| ELISA | Antibody-protein interaction | 1-5 ppm for commercial peanut kits [56] | Affected by protein denaturation during processing; may cross-react with related proteins [56] [50] | Directly measures allergenic proteins; high sensitivity; standardized protocols [1] | Antibody cross-reactivity; processing affects protein detectability [50] |
| LC-MS/MS | Mass detection of signature peptides | 1 mg/kg (pistachio) [50] | Complex matrices require optimized extraction; can detect multiple allergens simultaneously [74] [50] | High specificity and selectivity; multi-allergen detection; unaffected by antibody availability [50] | High equipment cost; requires skilled personnel; complex sample preparation [50] |
| PCR/qPCR | DNA amplification | ~1 ppm celery DNA in spiked samples [5] | Degraded DNA in processed foods requires short amplicons (200-300 bp) [5] [7] | DNA stability during processing; high specificity; can discriminate closely related species [5] [56] | Indirect detection; does not correlate directly with protein content; matrix inhibition [5] [50] |
| Biosensors | DNA hybridization or antibody-antigen binding | Varies by platform | Matrix effects can interfere with signal transduction; often requires extensive sample cleanup | Potential for rapid, on-site testing; portability; high-throughput capability [1] | Mostly in research phase; limited commercial availability; requires validation [1] |
Table 2: Impact of Food Processing on Detection Method Performance
| Processing Condition | Effect on Proteins | Effect on DNA | Recommended Method | Evidence |
|---|---|---|---|---|
| High-temperature processing (e.g., baking at 180-220°C) | Protein denaturation and modification; reduced antibody recognition [1] | DNA degradation; longer amplicons become undetectable [7] | DNA-based with short amplicons (<300 bp) or MS-based for modified proteins | Wheat and maize DNA detectability decreased with increasing baking temperature and time; primers for shorter fragments remained effective after 60min at 220°C [7] |
| Fermentation & Hydrolysis | Protein fragmentation; loss of conformational epitopes [75] | DNA largely unaffected | DNA-based methods | Protein-based methods may fail with hydrolyzed proteins, while DNA remains amplifiable [56] |
| Complex matrices (chocolate, spices, meat) | Matrix interference with antibody binding; sample cleanup critical [73] | PCR inhibition; requires DNA purification and quality assessment [5] | Protein-based with appropriate extraction or DNA-based with inhibition controls | xMAP FADA successfully detected allergens in challenging matrices like dark chocolate and sausages; DNA-based kits showed variable performance across product groups [5] [73] |
Experimental Protocols for Method Validation
Protein Extraction Protocol for Complex Matrices
Efficient protein extraction is fundamental for accurate allergen detection in protein-based methods. An optimized protocol achieves approximately 80% extraction efficiency across diverse food matrices [74]. The procedure begins with sample homogenization, followed by suspension in a buffered detergent solution (e.g., PBS with 0.05% Tween-20) for soluble proteins. For difficult-to-extract proteins, a reduction-denaturation step using SDS and β-mercaptoethanol improves recovery [73]. After incubation with continuous mixing, centrifuged supernatants are collected for analysis. For mass spectrometry applications, extracted proteins undergo enzymatic digestion (typically with trypsin) to generate peptides for LC-MS/MS analysis [50]. This protocol's effectiveness has been demonstrated in various matrices, including plant-based meat alternatives, with higher extraction efficiency correlating directly with improved identification reproducibility and more accurate allergen quantification [74].
DNA-Based Detection of Celery Allergens in Various Food Matrices
A comparative study evaluated three commercial DNA-based test kits for detecting celery in five food product groups representing different sectors of the AOAC food-matrix triangle [5]. The experimental protocol involved:
Sample Preparation: Blank food products (plant-based meat products, snacks, sauces, dried herbs and spices, and smoothies) were spiked with defined levels of characterized celery material (stem, root, greens, or seeds).
DNA Extraction: DNA was extracted from 100 mg of each sample using the CTAB-based method with additional purification steps. DNA quantity and quality were assessed using spectrophotometry (NanoDrop) [5] [7].
qPCR Analysis: Samples were analyzed in duplicate using celery-specific primers targeting the malate dehydrogenase gene. The qPCR reaction consisted of 5 μL of diluted DNA template (10 ng/μL) added to 20 μL of reaction mix containing TaqMan Universal Master Mix and specific primers/probe [5].
Data Analysis: Results were evaluated based on detection capability, quantification performance, and matrix effects. All kits performed according to specifications but showed clear matrix effects, with quantification proving challenging across all food product groups [5].
Method Selection Framework
Decision Pathway for Allergen Detection Method Selection
The following diagram illustrates the logical decision process for selecting the most appropriate allergen detection method based on testing objectives and food matrix characteristics:
Experimental Workflow for DNA-Based Allergen Detection
The standard workflow for DNA-based allergen detection in processed foods involves multiple critical steps to ensure accurate results:
Essential Research Reagents and Materials
Table 3: Research Reagent Solutions for Allergen Detection
| Reagent/Material | Function | Application Examples |
|---|---|---|
| CTAB Buffer | DNA extraction and purification; removes polysaccharides and polyphenols | DNA extraction from wheat, maize, and celery in processed foods [5] [7] |
| Proteinase K | Protein digestion; releases DNA from cellular structures | DNA extraction in Maxwell RSC PureFood GMO and Authentication Kit [5] |
| TaqMan Probes | Fluorescently-labeled probes for specific target detection in real-time PCR | Celery malate dehydrogenase gene detection with FAM-labeled probes [5] |
| Specific Antibodies | Recognition and binding to target allergenic proteins | xMAP FADA uses 29 antibodies conjugated to color-coded beads for 15 allergens [73] |
| Trypsin | Proteolytic enzyme for protein digestion into peptides for MS analysis | Digestion of pistachio and cashew proteins for LC-MS/MS analysis [50] |
| Isotopically Labeled Peptides | Internal standards for accurate quantification in mass spectrometry | Quantification of allergenic peptides in LC-MS/MS methods [50] |
The selection between protein-based and DNA-based allergen detection methods requires careful consideration of the specific analytical requirements and food matrix characteristics. Protein-based methods, particularly ELISA and LC-MS/MS, directly measure allergenic proteins and are preferred for compliance testing where regulatory thresholds are based on protein content. DNA-based methods offer advantages for processed foods where DNA stability exceeds that of proteins, and for discriminating between closely related species where antibody cross-reactivity may compromise protein-based assays.
Future methodological developments will likely focus on multiplex platforms that simultaneously detect multiple allergens, portable biosensors for rapid on-site screening, and improved extraction techniques that enhance recovery from complex matrices. The integration of computational tools for allergen prediction, such as AllergyPred [72] and iAller [76], may further complement analytical methods by providing preliminary risk assessments of novel proteins. As the food industry continues to evolve with alternative proteins and innovative processing technologies, allergen detection methods must similarly advance to ensure consumer protection and regulatory compliance.
Validation Criteria and the Role of Reference Materials in Standardization
Food allergies represent a significant global public health concern, affecting millions of individuals and requiring stringent food safety measures. The accurate detection of allergenic ingredients in food products is paramount for protecting sensitized consumers from potentially life-threatening reactions. This comparative guide examines the two predominant technological approaches for allergen detection: protein-based methods and DNA-based methods. Each methodology offers distinct advantages and limitations, with their reliability fundamentally dependent on robust validation criteria and well-characterized reference materials [8] [77]. The standardization of these analytical approaches ensures consistent results across different laboratories and commercial test kits, ultimately supporting regulatory compliance and consumer safety in the food industry [78] [79].
Core Detection Methodologies: A Comparative Framework
Protein-Based Detection Methods
Protein-based methods directly target the allergenic proteins themselves, which are the actual molecules that trigger immune responses in sensitized individuals.
- Enzyme-Linked Immunosorbent Assay (ELISA): This widely used immunological technique employs antibodies that specifically bind to allergenic proteins. It is considered the gold standard for quantitative allergen analysis in many applications due to its sensitivity and specificity [77] [79]. For instance, ELISA kits targeting casein and β-lactoglobulin are recommended for milk allergen detection [79].
- Mass Spectrometry (MS): LC-MS/MS methods identify and quantify unique peptide sequences derived from allergenic proteins. This approach offers high specificity and is increasingly used for multiplexed analysis of multiple allergens simultaneously [77] [79]. Mass spectrometry is particularly valuable for its ability to detect specific protein markers, with studies demonstrating its application in seafood allergen analysis by targeting parvalbumins [80].
- Multiplex Array Technology: Platforms like the Luminex xMAP system enable the simultaneous quantitative measurement of multiple specific food allergen proteins in a single assay using monoclonal antibodies and purified allergen reference standards [81].
DNA-Based Detection Methods
DNA-based methods indirectly detect the presence of allergenic ingredients by identifying species-specific DNA sequences.
- Real-Time Polymerase Chain Reaction (qPCR): This method amplifies and detects specific DNA sequences unique to the allergenic source. It provides high specificity for species identification and is less affected by food processing that may denature proteins [82] [83]. For example, qPCR has been successfully validated for detecting peanut traces with limits of detection as low as 0.5 ppm in various food matrices [83].
- Loop-Mediated Isothermal Amplification (LAMP): An isothermal DNA amplification technique that offers rapid results without requiring sophisticated thermal cycling equipment. When combined with lateral flow dipsticks (LFD), it provides a simple detection tool with sensitivity comparable to qPCR for targets like soybean [77].
Table 1: Core Methodologies for Food Allergen Detection
| Method Category | Specific Technology | Primary Target | Key Advantage | Major Limitation |
|---|---|---|---|---|
| Protein-Based | ELISA (Immunoassay) | Protein epitopes | High sensitivity, direct measurement of allergenic component | Antibody cross-reactivity, protein denaturation during processing |
| Mass Spectrometry (LC-MS/MS) | Peptide sequences | High specificity, multiplexing capability | Lower sensitivity vs. ELISA, complex instrumentation | |
| Multiplex Array | Multiple protein analytes | Simultaneous multi-allergen detection | Requires specialized equipment and standardized antibodies | |
| DNA-Based | Real-Time PCR (qPCR) | Species-specific DNA sequences | High specificity, resistant to protein denaturation | Indirect measurement (does not detect protein directly) |
| LAMP-LFD | DNA sequences | Rapid, simple equipment requirements | Less established for all allergens |
Critical Validation Parameters for Allergen Detection Methods
The reliability of any allergen detection method depends on rigorous validation against standardized performance criteria. Key parameters include:
- Sensitivity and Specificity: Sensitivity refers to the lowest detectable amount of an allergen (Limit of Detection, LOD), while specificity indicates the method's ability to exclusively identify the target allergen without cross-reacting with other species or matrix components [83]. For instance, a validated qPCR method for peanut detection demonstrated 98% sensitivity and 100% specificity across multiple food matrices [83].
- Accuracy and Precision: Accuracy represents the closeness of agreement between the measured value and the true value, often assessed through recovery experiments. Precision refers to the agreement between a series of repeated measurements under stipulated conditions [84] [82].
- Robustness and Repeatability: Robustness indicates the method's reliability under normal but variable operating conditions, while repeatability measures precision under identical conditions over a short time interval [83].
- Matrix Effects: Food matrices can significantly interfere with allergen detection. Validation must therefore assess method performance in relevant and often complex food backgrounds to ensure accurate quantification [84] [79].
Table 2: Key Validation Parameters and Performance Criteria
| Validation Parameter | Definition | Acceptance Criteria Examples | Impact on Method Reliability |
|---|---|---|---|
| Sensitivity (LOD) | Lowest detectable concentration | 0.5 ppm for peanut in grains (qPCR) [83] | Determines capability to detect trace-level contamination |
| Specificity | Ability to exclusively identify target | No cross-reactivity with almonds, soy, mustard, etc. [83] | Prevents false positives from cross-reactive species |
| Accuracy (Recovery) | Agreement between measured and true value | 80-120% recovery in spiked samples | Ensures quantitative results reflect actual allergen content |
| Repeatability | Precision under identical conditions | Consistent Ct values in qPCR duplicates [82] | Confirms method consistency within the same laboratory |
| Matrix Effects | Impact of food composition on detection | LOD of 2.5 ppm in dairy vs. 0.5 ppm in grains [83] | Highlights need for matrix-specific validation |
The Central Role of Reference Materials in Standardization
Reference materials (RMs) and certified reference materials (CRMs) are indispensable tools for achieving standardized and comparable results across different methods and laboratories.
Functions and Types of Reference Materials
- Calibration and Method Development: RMs with known allergen concentrations serve as calibration standards for quantitative methods, enabling the conversion of instrument signals (e.g., optical density in ELISA, Ct values in qPCR) to meaningful quantitative data [78] [79].
- Method Validation and Quality Control: These materials are essential for determining key validation parameters such as accuracy, precision, and limit of detection during method development and verification [79].
- Method Harmonization: Well-characterized RMs allow different laboratories to benchmark their results against a common standard, facilitating result comparability. For example, the National Institute of Standards and Technology (NIST) has developed tree nut reference materials (almond and hazelnut flours) to support consistent results for test kits used by regulators and food manufacturers [78].
Key Challenges and Current Gaps
Despite their critical importance, the allergen detection field faces significant challenges regarding reference materials:
- Limited Availability: There is a scarcity of certified reference materials, particularly for incurred allergens in processed food matrices [79].
- Material Characterization: Many existing RMs lack comprehensive characterization and assigned values for specific allergenic protein content, limiting their utility for quantitative analysis [78] [79].
- Processing Considerations: The detectability of allergens can be dramatically affected by food processing (e.g., heating, high-pressure treatment), which may alter protein structure or DNA extractability. This necessitates reference materials that reflect these processing-induced changes [84].
Experimental Workflows and Protocol Design
Protein-Based Workflow for Egg Allergen Detection
The following workflow is recommended for comprehensive egg allergen analysis in incident management scenarios:
Figure 1: Protein-based workflow for egg allergen detection. This multi-stage approach minimizes false negatives by employing orthogonal detection methods.
Detailed Protocol:
- Sample Preparation: A representative sample (100g-1kg) is homogenized into a powder or slurry using validated laboratory precautions to avoid cross-contamination [79].
- Protein Extraction: Allergenic proteins are extracted using appropriate buffers. The efficiency of extraction is critical, especially for processed foods where proteins may be denatured or bound to the matrix. Specialized extraction cocktails (e.g., the Méndez cocktail for gluten) containing reducing agents can improve recovery from processed matrices by breaking disulfide bonds [84].
- Initial ELISA Analysis: Test samples alongside suitable reference materials (e.g., NIST SRM 8445 for whole egg) using an ELISA kit targeting an egg white protein like ovalbumin. Include blank controls and spiked samples to monitor matrix effects and recovery [79].
- Secondary ELISA: If the initial test is negative, perform a second ELISA targeting a different egg white protein (e.g., ovomucoid) to account for potential variations in protein detectability [79].
- Confirmatory LC-MS/MS: If ELISA results remain negative, proceed with LC-MS/MS analysis targeting specific egg allergen peptides. The method developed during the EFSA ThRAll project provides the sensitivity required to quantify egg at action levels identified by international standards [79].
DNA-Based Workflow for Peanut Allergen Detection
The following workflow outlines the validation and application of qPCR for peanut detection:
Figure 2: DNA-based workflow for peanut allergen detection validation. This systematic approach ensures comprehensive method characterization.
Detailed Protocol:
- Matrix Selection and Spiking: Select five food matrix categories (bakery, meats, ready-to-eat, dairy, grains). Prepare blank samples confirmed peanut-free, then spike with peanut material at 1000 ppm. Create serial dilutions down to 0.5 ppm for sensitivity determination [83].
- DNA Extraction: Extract DNA using commercial kits (e.g., ION Force FAST). Include steps to remove potential PCR inhibitors present in complex food matrices. Assess DNA purity and concentration spectrophotometrically [83].
- qPCR Amplification: Perform real-time PCR using validated primer/probe systems. The SPECIALfinder MC Peanut kit targets specific peanut DNA sequences. Include inhibition controls to monitor PCR efficiency [83].
- Specificity Assessment: Test cross-reactivity by analyzing samples spiked with potentially cross-reactive species (almonds, soy, mustard, celery, peas, carrots, cocoa). No amplification should occur with these non-target species [83].
- Data Analysis and Validation: Calculate LOD (0.5 ppm for most matrices), LOQ, sensitivity (98%), and specificity (100%). Assess repeatability through multiple replicates and robustness by introducing minor variations in experimental conditions [83].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Research Reagents and Materials for Allergen Detection Studies
| Reagent/Material | Function | Examples/Specifications |
|---|---|---|
| Certified Reference Materials (CRMs) | Method calibration and validation | NIST RM 8404 (Almond Flour), NIST RM 8405 (Hazelnut Flour) [78] |
| Incurred Reference Materials | Assess impact of food processing on detectability | Egg in chocolate, milk in baked matrices [79] |
| Monoclonal/Polyclonal Antibodies | Target capture and detection in immunoassays | R5 antibody for gluten, anti-casein for milk, anti-parvalbumin for fish [77] [80] |
| Species-Specific Primers/Probes | DNA amplification in PCR-based methods | Chloroplast markers (matK) for peanut, 2S albumin gene for sesame [77] [83] |
| Specialized Extraction Buffers | Efficient recovery of allergens from complex matrices | Méndez cocktail for gluten (contains reducing agents) [84] |
| Quality Control Materials | Routine performance monitoring | Surplus proficiency testing materials, in-house prepared controls [79] |
The comparative analysis of protein-based and DNA-based allergen detection methods reveals that these approaches offer complementary, rather than competing, capabilities. Protein-based methods directly measure the allergenic component, making them ideal for quantitative risk assessment, while DNA-based methods provide exceptional specificity for ingredient identification. The reliability of both methodologies is fundamentally dependent on rigorous validation against standardized criteria and the availability of well-characterized reference materials. Future advancements in allergen detection will likely focus on multiplexed platforms that simultaneously target multiple allergens, the development of more sophisticated reference materials that reflect processing impacts, and the integration of orthogonal detection principles to minimize false results. As international regulatory frameworks continue to evolve, standardized validation protocols and harmonized reference materials will play an increasingly critical role in ensuring the accuracy and comparability of allergen detection data, ultimately enhancing consumer protection and supporting evidence-based food safety policies.
Conclusion
This analysis affirms that the choice between protein-based and DNA-based allergen detection is not a matter of superiority but of context. Protein-based methods, particularly ELISA and emerging mass spectrometry, are indispensable for directly quantifying the allergenic agent, essential for compliance with threshold-based regulations. DNA-based methods offer robust, sensitive, and specific detection, especially valuable for highly processed foods where protein structures are denatured. The future of allergen detection lies in integrating these technologies, leveraging the strengths of each, and embracing innovations such as biosensors, AI-powered diagnostics, and high-throughput multiplexed platforms. For biomedical research, these advancements will enable more precise risk assessment, support the development of novel therapeutics and hypoallergenic foods, and ultimately enhance clinical diagnostics and patient safety.
References
- 1. Research progress on detection methods for food allergens [https://www.sciencedirect.com/science/article/abs/pii/S0889157524009402]
- 2. Food Allergenicity Evaluation Methods: Classification, ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12191692/]
- 3. Advances in Food Processing Techniques for Allergenicity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12651889/]
- 4. The Growing Importance of Allergen Testing: How Certified ... [https://fsns.com/the-growing-importance-of-allergen-testing-how-certified-group-ensures-food-safety-with-elisa/]
- 5. Comparison of commercial DNA kits for allergen detection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11419853/]
- 6. Innovations shaping the future of food allergen testing [https://allergenbureau.net/innovations-shaping-the-future-of-food-allergen-testing/]
- 7. Development of PCR Methods for Detecting Wheat and ... [https://www.mdpi.com/2673-6284/14/4/78]
- 8. Detection of Allergenic Proteins in Foodstuffs: Advantages of the Innovative Multiplex Allergen Microarray-Based Immunoassay Compared to Conventional Methods - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8949930/]
- 9. Rapid Identification of soybean and wheat allergens using ... [https://www.sciencedirect.com/science/article/abs/pii/S2212429225020139]
- 10. Development of PCR Methods for Detecting Wheat and Maize ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12551070/]
- 11. Review of allergen analytical testing methodologies: Allergen detection methods: Unbiased literature search | Food Standards Agency [https://www.food.gov.uk/research/review-of-allergen-analytical-testing-methodologies-allergen-detection-methods-unbiased-literature-search]
- 12. Comparison of commercial DNA kits for allergen detection of celery in food matrices - ScienceDirect [https://www.sciencedirect.com/science/article/pii/S2405844024128551]
- 13. Dual-color blending based visual LAMP for food allergen ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9039889/]
- 14. The role of DNA-based biosensors in species identification ... [https://www.sciencedirect.com/science/article/pii/S0924224424000268]
- 15. DNA-Based Biosensors for the Biochemical Analysis [https://pmc.ncbi.nlm.nih.gov/articles/PMC8945906/]
- 16. Comparison of Detection Limits for Allergenic Foods ... [https://www.sciencedirect.com/science/article/pii/S0362028X22103376]
- 17. Pushing the detection limits: strategies towards highly ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8346249/]
- 18. Food Allergen Testing Market [https://www.futuremarketinsights.com/reports/food-allergen-testing-market]
- 19. How US Food Allergen Testing Works — In One Simple ... [https://www.linkedin.com/pulse/how-us-food-allergen-testing-works-one-simple-flow-r8dme/]
- 20. United States Food Allergen Testing Market Trends ... [https://vocal.media/feast/united-states-food-allergen-testing-market-trends-and-summary-2025-2033]
- 21. Comparison of commercial lateral flow immunoassays and ... [https://pubmed.ncbi.nlm.nih.gov/32659710/]
- 22. COMPARISON OF PERFORMANCE CHARACTERISTICS ... [https://www.medrxiv.org/content/10.1101/2021.04.29.21256260v1]
- 23. Lateral Flow Immunoassay Basics [https://www.jacksonimmuno.com/technical/products/applications/elisa/lateral-flow/immunoassays-introduction]
- 24. Targeted Proteomics: Current Status and Future ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5082697/]
- 25. Food allergen detection by mass spectrometry: the role of ... [https://www.nature.com/articles/npjsba201622]
- 26. Protein or No Protein? Opportunities for DNA-Based ... [https://pubmed.ncbi.nlm.nih.gov/30180550/]
- 27. DNA, protein and aptamer-based methods for seafood ... [https://pubmed.ncbi.nlm.nih.gov/34184960/]
- 28. BatMass [https://batmass.org/]
- 29. Visual mass-spec share (vMS-Share): a new public web- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6605085/]
- 30. Why is Real-time PCR is superior to end-point PCR? [https://help.medicinalgenomics.com/en/qpcr-vs-end-point-pcr]
- 31. Comparison of real-time polymerase chain reaction and end-point polymerase chain reaction for the analysis of gene expression in preimplantation embryos - PubMed [https://pubmed.ncbi.nlm.nih.gov/16554012/]
- 32. How To Interpret RT-qPCR Results [https://www.goldbio.com/blogs/articles/how-to-interpret-rt-qpcr-results?srsltid=AfmBOoqbYWc5gZg_EPAN-yxoGFuW91CQcSLUSmGng7WKUrYrdcHz0TuV]
- 33. Multiplex Polymerase Chain Reaction - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/multiplex-polymerase-chain-reaction]
- 34. 2048. Comparison Between Endpoint and Real-Time (RT) Polymerase Chain Reaction (PCR) for the Diagnosis of Pneumocystis Pneumonia (PCP) - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6253075/]
- 35. Statistical analysis of real-time PCR data - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC1395339/]
- 36. The application of microfluidic technology in allergen ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12150919/]
- 37. Recognition Element-Based Strategies for Rapid Detection ... [https://www.mdpi.com/2079-6374/15/11/717]
- 38. Progress in the application of isothermal amplification ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12271116/]
- 39. Microfluidic chip and isothermal amplification technologies for ... [https://jbioleng.biomedcentral.com/articles/10.1186/s13036-022-00312-w]
- 40. Real-time amplification and high resolution melt analysis ... [https://www.sciencedirect.com/science/article/abs/pii/S0003267025004404]
- 41. LAMP-Based 4-Channel Microfluidic Chip for POCT ... [https://www.mdpi.com/2079-6374/15/8/506]
- 42. An extraction-free and one-pot two-temperature CRISPR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12506078/]
- 43. Nucleic acid amplification tests in digital microfluidics [https://www.nature.com/articles/s41378-025-00977-5]
- 44. Real-time microfluidic RPA chip for simultaneous ... [https://www.sciencedirect.com/science/article/abs/pii/S088915752501066X]
- 45. Rapid and accurate electrochemical sensor for food ... [https://www.nature.com/articles/s41598-021-00241-6]
- 46. From Matrix to Protein: The Evolving Approach to Allergen ... [https://www.romerlabs.com/en/library/knowledge/detail/from-matrix-to-protein-the-evolving-approach-to-allergen-quantification-in-food]
- 47. The Importance of Food Matrix in Diet Quality and Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10201811/]
- 48. Why are scientists so intrigued by the food matrix? [https://www.bbc.co.uk/food/articles/food_matrix]
- 49. Food allergen testing: should you use ELISA, PCR, or LFA ... [https://blog.invitek.com/articles/foodsafetyintegrity/food-allergen-testing-should-you-use-elisa-pcr-or-lfa-methods]
- 50. Allergens in Food: Analytical LC-MS/MS Method for the ... [https://www.mdpi.com/2304-8158/14/17/3031]
- 51. False Positivity of Anti-SARS-CoV-2 Antibodies in Patients ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9320684/]
- 52. False Positive Results With SARS-CoV-2 RT-PCR Tests ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7934325/]
- 53. False Positives in PCR Tests for COVID-19 - ICD10monitor [https://icd10monitor.medlearn.com/false-positives-in-pcr-tests-for-covid-19/]
- 54. Differential patterns of cross-reactive antibody response ... [https://www.nature.com/articles/s41598-022-20849-6]
- 55. Rare but persistent false positives on COVID-19 home ... [https://www.umassmed.edu/news/news-archives/2024/03/rare-but-persistent-false-positives-on-covid-19-home-antigen-tests-reported-in-nejm-letter-by-umass-chan-researchers/]
- 56. Advanced DNA-based methods for the detection of peanut ... [https://www.sciencedirect.com/science/article/abs/pii/S0165993618305648]
- 57. Optimising Extraction of Specific Food Allergens from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12563046/]
- 58. An integrated microfluidic platform for on-site qPCR analysis [https://pubs.rsc.org/en/content/articlehtml/2025/lc/d4lc00570h]
- 59. New Research in Food Allergen Detection [https://www.mdpi.com/2304-8158/11/10/1520]
- 60. First nanoplate digital PCR method to trace allergenic foods [https://www.sciencedirect.com/science/article/pii/S030881462400298X]
- 61. Reference Gene-Assisted LAMP–LFD for Sensitive and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12063182/]
- 62. Current Trends in Proteomic Advances for Food Allergen ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7563520/]
- 63. Enzyme- and nanozyme-based food allergen detections [https://www.sciencedirect.com/science/article/pii/S0924224425001529]
- 64. A generalizable decision-making framework for selecting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10987262/]
- 65. Cost Comparisons of Physician-Ordered Versus Direct-to- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11663041/]
- 66. Health Care Cost Institute: Employers in Seven States Pay ... [https://www.darkdaily.com/2023/06/14/health-care-cost-institute-employers-in-seven-states-pay-up-to-6x-more-for-hospital-lab-tests-as-compared-to-same-tests-in-doctor-offices-and-independent-labs/]
- 67. Comparative cost analysis of point-of-care versus laboratory ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0223669]
- 68. Onsite Vs Offsite Lab Services Which Is Right for Your ... [https://nightfallmedical.com/onsite-lab-services-vs-offsite-lab-services-which-is-right-for-your-practice/]
- 69. On-Site vs. Testing Facility Material Testing [https://prycoglobal.com/blog/on-site-vs-testing-facility-where-should-material-testing-occur/]
- 70. Central Lab vs. Local Lab in Clinical Trials [https://3billion.io/blog/central-lab-vs-local-lab-in-clinical-trials]
- 71. Lab Testing vs. Real-World Testing: Understanding the ... [https://valuetransform.com/lab-testing-vs-real-world-testing-understanding-the-differences/]
- 72. AllergyPred: a web server for allergen prediction - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12230706/]
- 73. Multi-laboratory validation of the xMAP—Food Allergen ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7347184/]
- 74. Efficient protein extraction for assessing food allergy risk in ... [https://www.sciencedirect.com/science/article/pii/S0308814624028711]
- 75. Review of Methods for the Detection of Allergens in Novel ... [https://science.food.gov.uk/article/125903-review-of-methods-for-the-detection-of-allergens-in-novel-food-alternative-proteins]
- 76. Computational prediction of allergenic proteins based on ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2023.1294159/full]
- 77. New Research in Food Allergen Detection - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9141938/]
- 78. NIST Tree Nut Reference Materials Support Food Allergen ... [https://www.nist.gov/news-events/news/2022/06/nist-tree-nut-reference-materials-support-food-allergen-testing]
- 79. Review of allergen analytical testing methodologies [https://www.food.gov.uk/research/review-of-allergen-analytical-testing-methodologies-allergen-testing-workflows-to-support-incident-management]
- 80. Protein and DNA-based assays as complementary tools for fish allergen detection* - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6040006/]
- 81. Standardization of Food Allergen Measurements Using ... [https://pubmed.ncbi.nlm.nih.gov/37737988/]
- 82. Comparative assessment of DNA-based approaches for ... [https://www.sciencedirect.com/science/article/abs/pii/S0308814614004683]
- 83. Detection of Peanut Traces in Food by an Official Food Safety ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8909111/]
- 84. Use of incurred samples to determine the performance ... [https://allergenbureau.net/use-of-incurred-samples-to-determine-the-performance-of-allergen-detection-methods/]